Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 09 February 2023

Recent advances in plant disease severity assessment using convolutional neural networks
- Tingting Shi 1 , 2 ,
- Yongmin Liu 1 , 2 ,
- Xinying Zheng 3 ,
- Kui Hu 1 , 2 ,
- Hao Huang 1 , 2 ,
- Hanlin Liu 1 , 2 &
- Hongxu Huang 1 , 2
Scientific Reports volume 13 , Article number: 2336 ( 2023 ) Cite this article
8890 Accesses
18 Citations
Metrics details
- Computer science
- Plant biotechnology
In modern agricultural production, the severity of diseases is an important factor that directly affects the yield and quality of plants. In order to effectively monitor and control the entire production process of plants, not only the type of disease, but also the severity of the disease must be clarified. In recent years, deep learning for plant disease species identification has been widely used. In particular, the application of convolutional neural network (CNN) to plant disease images has made breakthrough progress. However, there are relatively few studies on disease severity assessment. The group first traced the prevailing views of existing disease researchers to provide criteria for grading the severity of plant diseases. Then, depending on the network architecture, this study outlined 16 studies on CNN-based plant disease severity assessment in terms of classical CNN frameworks, improved CNN architectures and CNN-based segmentation networks, and provided a detailed comparative analysis of the advantages and disadvantages of each. Common methods for acquiring datasets and performance evaluation metrics for CNN models were investigated. Finally, this study discussed the major challenges faced by CNN-based plant disease severity assessment methods in practical applications, and provided feasible research ideas and possible solutions to address these challenges.
Similar content being viewed by others

Construction of deep learning-based disease detection model in plants

Advanced deep learning techniques for early disease prediction in cauliflower plants

CPD-CCNN: classification of pepper disease using a concatenation of convolutional neural network models
Introduction.
Plant diseases caused by various organisms that damage plant growth, such as pests, bacteria or fungi, are a major cause of agricultural losses. Reliable and accurate methods for assessing disease severity are essential for effective disease control and minimizing yield loss 1 . There are several ways to assess the severity of plant diseases. The traditional method of determining disease severity is by Visual Assessment, which is highly unreliable due to the similarity of diseases and the diversity of characteristics that are susceptible to external factors and subjective individual differences. Visual Assessment usually needs to be carried out by experienced specialists, which is not efficient, and many farmers do not have access to specialists, making accurate and timely disease severity identification very difficult. In addition, hyperspectral imaging has been used to measure the severity of plant diseases, but this technique requires sophisticated equipment such as sensors and a certain level of expertise, making it costly and inefficient 2 .
In recent years, with the rapid development of computer imaging technology and the continuous improvement of the hardware performance of related electronic devices, computer vision and artificial intelligence (AI) have been widely used in the field of agricultural diagnosis, such as plant species classification, leaf disease identification and plant disease severity estimation 3 . Deep learning has now made significant breakthroughs in the field of computer vision, and CNN has shown excellent performance in plant disease detection applications. Compared to traditional methods, CNN is able to automatically and directly extract features from the input image, eliminating the need for complex image pre-processing and enabling end-to-end detection methods 4 . To date, satisfactory results have been achieved in identifying plant disease species using CNN, but little research has been done in the area of disease severity assessment. This study focuses on the application of CNN for plant disease severity assessment, and systematically reviews the related research to provide reference ideas for further research work.
The remainder of this review is organized as follows: the second part provides an overview of the concepts related to the Visual Assessment of plant disease severity. The third part reviews the history of the development of CNN. The fourth part deals with the specific application of CNN on plant disease severity, illustrating the differences between single-task and multi-task systems. And we focus on the basic working principles of CNN-based plant disease severity assessment methods from three aspects: classical CNN framework, improved CNN architecture, and CNN-based semantic segmentation network, and analyze the advantages and disadvantages of each method. The fifth part summarizes the relevant public datasets and presents CNN performance evaluation metrics. The sixth section discusses the major challenges that CNN-based plant disease severity assessment may face in practical applications, and provides feasible research ideas and possible solutions to these challenges.
visual assessments
Definition of plant disease severity.
Plant disease severity, defined as the ratio of plant units with visible disease symptoms to the total plant unit (e.g. leaves), is an important quantitative indicator for many diseases 5 . Timely and accurate assessment of disease severity is critical in crop production because disease severity directly affects crop yield and is often used as a predictor to estimate crop loss with excellent accuracy 6 . For example, severity indicators can be used as decision thresholds or disease forecasts to help growers rationalize disease control, such as deciding on the dose and type of pesticide and the time of day to spray.
Visual assessment methods for plant disease severity
Accurate measurement and evaluation of disease severity is critical to agricultural production. Because it ensures a correct analysis of treatment effects, an accurate understanding of the correlation between yield loss and disease severity, and a reasonable assessment of plant growth stages 7 . Inaccurate or unreliable disease assessment can lead to erroneous conclusions, resulting in the wrong disease management actions, which can further exacerbate losses. Assessment of disease severity is typically done using a variety of scales, including nominal (descriptive) scales, ordinal rating scales, interval (category) scales, and ratio scales 1 . The following is an overview of these scales for visual assessment of disease severity, both qualitative and quantitative.
Qualitative scales.
Descriptive scale: This is one of the simplest and most subjective criteria in the disease severity grading scales. The disease is divided into several categories with descriptive terms such as mild, moderate, and severe. Due to the subjectivity and lack of quantitative definitions, the value of this scale is very limited, except for ratings in a specific condition.
Qualitative ordinal scale: This is still the descriptive disease scale, but provides more variety in the categories of disease severity levels than the descriptive scale. For example, Xu et al. 8 assigned a scale of 0–5 to describe the severity of symptoms of zucchini yellow mosaic virus and watermelon mosaic virus to indicate increasing disease severity. This scale is widely used for certain diseases, especially for assessing viral diseases with symptoms that are not easily quantifiable 1 , 5 .
Quantitative scales.
Quantitative ordinal scale: This scale consists of numbers in known categories, usually the percentage of symptomatic areas. It can be further divided into two types: equal interval and unequal interval. However, equal interval rating scales may give a higher average severity, especially if the actual severity is at the lower end of a category, because the interval is so wide that it is difficult to show differences, leading to an inaccurate rating 9 . Some disease rating scales have unequal intervals. The Horsfall-Barratt scale (H–B scale) is a widely used unequal interval scale. It was developed by Horsfall and Barratt 10 , which effectively alleviates the problem of equal intervals. For example, Bock et al. 11 used the scale to estimate the severity of citrus ulcer disease. Forbes et al. 12 used the H-B scale to estimate the severity of potato late blight in the field, etc.
Ratio scale: This scale is widely used for visual assessment of severity. The grader measures the percentage of symptomatic organs, defined as 0% to 100%, and rates the severity accordingly. Therefore, the ratio scale places greater demands on the rater to identify and measure the actual disease more accurately.
Although plant disease severity can be assessed by several different methods, both qualitative and quantitative assessment methods tend to result in assessments that are inconsistent with reality due to factors such as the subjectivity of individual raters, the tendency to overestimate disease severity when it is low, and the bias of raters toward 5% whole number intervals 13 . To improve the accuracy of rater estimates, the Standard Area Map (SAD) has long been used as a tool to help estimate plant disease severity 14 , 15 . Professional training of raters can also be effective in improving the accuracy of the assessment.
History of CNN development
Deep learning began with the introduction of threshold logic in 1943 and is essentially a process of building computer models that closely resemble human neural networks 16 . CNN is a subset of deep learning that first appeared in the 1980s 17 . In the beginning, the concept of receptive field was developed and later introduced into CNN research 18 . Later, with the introduction of the BackPropagation (BP) algorithm and the training of multi-layer perceptron, researchers tried to automatically extract features instead of manually designing features 19 . LeCun et al. 20 proposed a CNN architecture called “LeNet-5” using BP networks, which outperformed all other techniques on a standard handwritten digit recognition task at the time. Research on deep neural network models was put on hold due to a number of problems encountered with traditional BP neural networks, such as local optima, overfitting, and gradient disappearance with increasing number of network layers, and the accompanying proposal of some shallow machine models at that time 19 . Until about 2006, Hinton et al. 21 found that artificial neural networks with multiple hidden layers have excellent feature learning capabilities. Glorot et al. 22 mitigated the problem of disappearing gradients during training with a normalization method. Attention shifted back to deep learning. In 2012, AlexNet 23 won the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC), and since then, DL has attracted the attention of more and more researchers, and AlexNet is considered a major breakthrough in the field of deep learning. Next, the CNN architecture continues to evolve, and many algorithms with excellent performance emerge. The main classical CNN networks are LeNet, AlexNet, VGG 24 , GoogLeNet 25 , Resnet 26 , DenseNet 27 , and so on. The evolutionary sequence order from LeNet to DenseNet is shown in Fig. 1 .
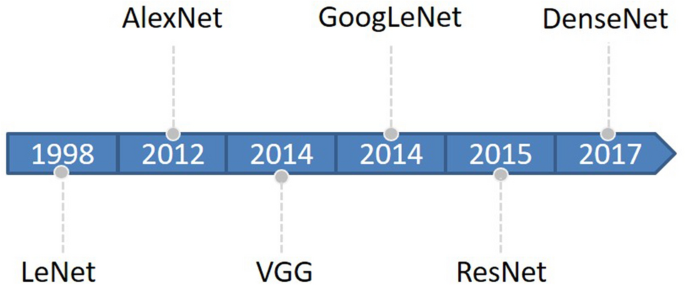
Evolution timeline of CNNs from LeNet to DenseNet.
As CNN evolves, new CNN models are constantly emerging that implement different features. For example, lightweight networks: SqueezeNet 28 , MobileNet 29 , ShuffleNet 30 , Xception 31 , EfficientNet 32 . Target detection networks: R-CNN 33 , Fast R-CNN 34 , Faster R-CNN 35 , YOLO 36 , SSD 37 . Segmentation networks: FCN 38 , SegNet 39 , U-Net 40 , PSPNet 41 , DeepLab 42 , Mask RCNN 43 , etc., they show excellent performance and great research value.
CNN-based plant disease severity assessment method
CNN has been used with great success to assess the severity of plant diseases. Automatic estimation of plant disease severity based on CNN was first proposed by Wang et al. 44 in 2017. They used different CNN models to classify apple black rot images with four severity levels and achieved an overall accuracy of 90.4% on the test set, suggesting that CNN is a promising new technique for fully automated plant disease severity classification. Liang et al. 3 proposed PD 2 SE-Net to implement a multitask system for disease severity estimation, plant species identification, and plant disease classification with overall accuracies of 0.91%, 0.99%, and 0.98%, respectively. Su et al. 45 combined ResNet-101 network and semantic segmentation to rapidly predict the severity of Fusarium head blight (FHB) in wheat with a prediction accuracy of 77.19%.
Single-task versus multi-task systems
Deep learning tends to focus on optimizing for specific metrics. In other words, a model or a set of models is often trained to perform the single target task, and such systems are known as single-task systems 46 . On the other hand, there is the concept of multi-task learning (MTL), where multiple tasks can be learned simultaneously if they are linked together 47 . Experimental studies have shown that learning features from multiple related tasks simultaneously is more beneficial than learning them independently in terms of prediction performance. MTL can reduce the risk of overfitting in each task by learning tasks in parallel and thus using more features from different tasks, leading to better generalization of the model 48 , 49 , 50 .
Studies using CNN for plant disease detection include single-task systems that individually identify plant disease species or estimate disease severity. For example, Prabhakar et al. 51 used ResNet101 to assess the severity of leaf blight in tomato. Zeng et al. 52 trained six different CNN models to classify the severity of citrus yellow shoot. There are also multitasking systems that perform both tasks simultaneously. For example, José G.M. Esgario et al. 46 used CNN to implement a classification of coffee leaf disease species and severity grading. Fenu et al. 53 considered five pre-trained CNN architectures as feature extractors for the classification of three diseases and six severity levels, whose experimental results show that the trained model is robust in automatically extracting disease leaf identification features using a multi-task learning model.
Application of CNN in plant disease severity assessment
To clarify the specific implementation process of CNN for plant disease severity assessment, 16 high-quality articles that fit the research topic were selected for this study. First, a search was conducted on the Web of Science platform, one of the world's largest and most comprehensive scientific information resources 54 . According to 55 , the process of collecting research sets requires the definition of search terms, so the keywords of “Convolutional neural network” (Topic) and “plant disease severity” (Topic) were entered into Web of Science, and as of 2022, 57 articles were retrieved with the year of publication shown in Fig. 2 . Among the 57 papers, 16 papers were selected for specific analysis based on the research object (plant disease) and research method (CNN). On this basis, the most recent research in 2022 is analyzed separately. According to the different CNN network architectures used in these 16 articles, they are further divided into three categories: classical CNN framework, improved CNN architecture, and CNN-based segmentation network. The flowchart of the CNN-based method for plant disease severity assessment method is shown in Fig. 3 .
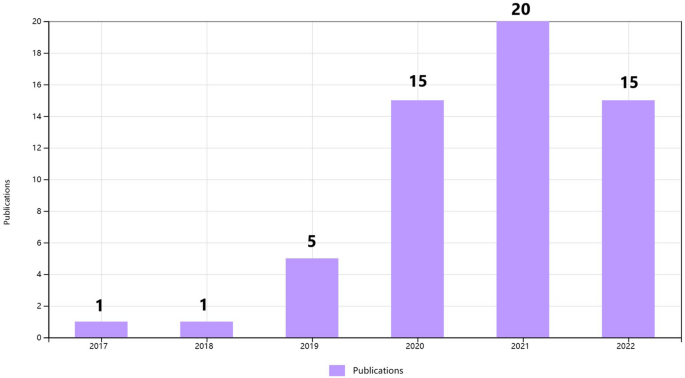
The distribution graph of the publication years of 57 articles based on the keywords of “convolutional neural network” and “plant disease severity” (from Web of Science).
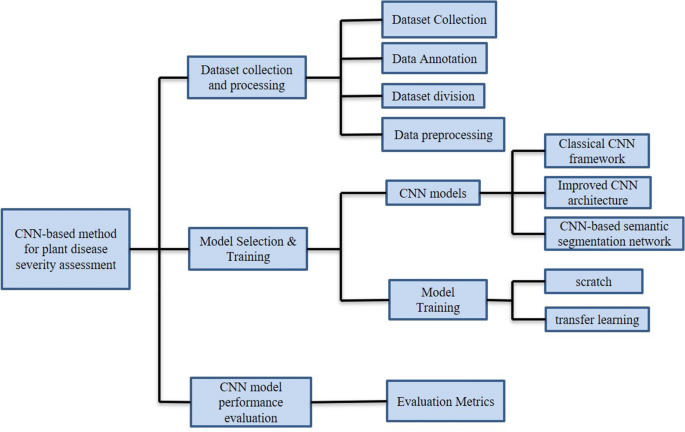
The flowchart of the CNN-based plant disease severity assessment method.
Classical CNN framework
10 of the 16 articles are based on the classical CNN framework for implementing severity grading. The 10 studies differ in specific CNN frameworks and research topics, but are similar in that they are CNN-based approaches to assessing plant disease severity. Therefore, they have similarities in the specific implementation process. The process of implementing plant disease severity assessment based on the classical CNN framework can be divided into the following three main steps.
The first step is to collect and process datasets, and is described in four aspects.
Dataset characteristics. Of the 10 studies, 6 studies used self-made datasets and 4 studies used the images from PlantVillage images. The self-made datasets can be further divided into two types. One is for images taken under controlled conditions. For example, in 46 , The photos were taken from the abaxial (lower) side of the leaves under partially controlled conditions and placed on a white background. The other is for images taken under natural conditions with a complex background. In contrast, the background of the images in PlantVillage is uniform and homogeneous. Making your own dataset is a time-consuming and expensive process, but it is more in line with what happens in a real environment. A large number of studies have demonstrated that when models trained on controlled images are used to predict images collected from real-world environments, their accuracy is significantly reduced 56 , 57 , 58 . If a public dataset does not meet the needs of a particular study, self-made datasets must be produced.
Dataset annotation. One of the necessary conditions for assessing severity is that the records are labeled with different severity levels. Of the 10 articles, 3 were labeled according to the descriptive scale, 1 according to the qualitative ordinal scale, 4 according to the quantitative ordinal scale, 1 article did not indicate the labeling method in the article. For example, in 46 , a quantitative ordinal scale was used. Severity was classified into five levels according to the proportion of diseased leaves: healthy (< 0.1%), very low (0.1–5%), low (5.1–10%), high (10.1%-15%), and very high (> 15%).
Dataset division. The dataset is usually divided into three parts: training dataset, validation dataset and test dataset. The training set is used to train the model, the validation set is used to tune the hyperparameters, and the test set is used to evaluate the model performance 59 . The 10 studies all basically used 70% to 85% of the dataset for training. Mohanty et al. 60 tried five different separation ratios to partition the dataset, and the experimental results showed that using 80% of the dataset for training and 20% for validation was ideal for their data.
Data preprocessing. Typically, two preprocessing operations are performed before the images are fed into the CNN. One is to resize the images to match the input layer requirements. For example, The image size in PlantVillage is 256 × 256, and the AlexNet input layer requires a size of 227 × 227, then the original photos need to be resized. This processing is reflected in all 10 studies. Second, the images are normalized to help the model converge faster, significantly improving the efficiency of end-to-end training 61 .
The second step is the model selection and training phase, which is described below in two aspects.
CNN framework selection. The CNN frameworks used in the 10 studies include AlexNet, VGG, GoogLeNet, ResNet, DenseNet, MobileNet, Inception, Faster R-CNN, YOLO, EfficientNet, SqueezeNet, Xception, etc. The vast majority of these studies have used multiple CNN frameworks in comparative experiments to determine which model is better at detecting the severity of a particular plant disease under the same training conditions 52 .
Training methods. There are two ways to train CNN, one is to start from scratch and the other is transfer learning. Transfer learning refers to adapting a pre-trained network on a large set of images, such as ImageNet (1.2 million images in 1000 classes), to a different task, which is implemented by the underlying CNN learning non-specific features 62 . There are two approaches to transfer learning: feature extraction and fine-tuning. Feature extraction is the process of keeping the weights of a pre-trained model unchanged and then using them to train a new classifier on the target dataset. Fine-tuning involves initializing the model using the weights from a pre-trained model, and then training some or all of the weights on the target dataset 63 . Brahimi et al. 64 used three approaches of feature extraction, fine-tuning, and training from scratch to train six CNN models. And the results suggested that the fine-tuning models had the highest accuracy and the feature extraction models had the shortest training time. 8 of the 10 studies used transfer learning and only one was trained from scratch. In 44 , the two methods of training models were compared and the results showed that transfer learning alleviated the problem of insufficient training data.
The final step is to evaluate the performance of the CNN models.
The performance of CNN models is obtained by using the test set on the trained model. It is critical that the test set is independent of the training and validation sets, otherwise the evaluation results may be highly biased. Mohanty et al. 60 trained a model to identify 14 crops and 26 diseases with an overall accuracy of 99.35% on a test set, where there was no clear separation between the validation and test set. When they tested the model on a set of images taken under different conditions than the training images, the model’s accuracy dropped dramatically to 31%. It is worth noting that only 4 of the 10 studies explicitly distinguish three types of datasets. Sibiya et al. 65 explicitly separated the training set, validation set, and test set. The experimental results showed that the proposed model was neither overfitting nor underfitting, as the model achieved the accuracy of 95.63% on the validation set and a high accuracy of 89% on the test set.
Quantitative assessment of model performance is achieved through evaluation metrics. Evaluation metrics typically include accuracy, precision, recall, mean average precision (mAP), and F1 score based on precision and recall. With the development of deep learning, the performance of CNN models on different datasets has been improved, and various evaluation metrics have been increased. A consistent performance comparison of CNNs from different studies is difficult to achieve because most CNN-based studies for plant disease severity assessment apply to specific datasets, many of which are not yet publicly available and do not provide all the parameters needed to reproduce experiments.
Improved CNN architecture
2 of the 16 articles are based on an improved CNN architecture for severity assessment. Comparing the classical CNN framework with the improved CNN architecture, the similarity is that the implementation process is basically the same, and the difference is that the latter uses an improved network based on the classical CNN with the aim of designing a higher performance and more practical system for plant disease diagnosis.
In 3 , a network, PD 2 SE-Net, was proposed to design a more excellent and practical plant disease diagnosis system. PD 2 SE-Net introduced the ResNet50 network as the base model and integrated the building blocks of ShuffleNet-V2 30 . The PD2SE-Net architecture is shown in Fig. 4 . There are two key components of the PD 2 SE-Net that make it so effective. One is the introduction of a residual structure to construct the parameter sharing layers, which allows the model more information to update per batch. Inspired by ShaResNet 66 , ResNet50 was used to build the basic framework and integrated with parameter sharing to reduce the redundant information in the network. The other is the introduction of shuffle units. The ShuffleNet-V2 units were used to extract the feature maps of different plant species and diseases with low computational complexity. Finally, PD 2 SE-Net achieved plant species recognition, disease classification, and severity estimation with overall accuracies of 0.99, 0.98, and 0.91, respectively.
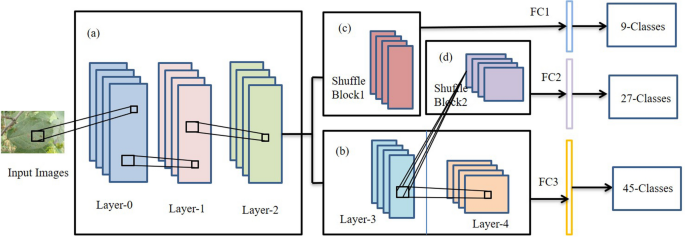
The architecture of PD2SE-Net. Five parts: ( a ) parameter sharing layer; ( b ) the third layer is the parameter sharing layer between the fourth layer and shuffling block 2, and the fourth layer is the high-dimensional feature extractor for severity estimation; ( c ) a feature extractor for plant species recognition; ( d ) feature extractor for plant disease diagnosis; ( e ) fully connected layers 3 .
Xiang et al. 67 proposed a lightweight network, L-CSMS, based on residual networks, channel shuffle operation and multi-size module for plant disease severity assessment. Multiple convolution kernels of different sizes were used in the multiscale convolution module to extract different receptive fields in order to obtain robust features and spatial relationships from feature maps 68 . Channel shuffle operation was introduced to enable information communication between different channel groups and to improve accuracy. The channel shuffle operation and the multi-size convolution module were integrated into the building block as a stacked topology, as is shown in Fig. 5 . L-CSMS used the residual learning approach of ResNet to build a deep network by stacking modules of the same topology. To validate the performance of the L-CSMS model, Xiang et al. 67 conducted comparative experiments between the L-CSMS model and ResNet, DenseNet, Inception-V4, PD 2 SE-Net, ShuffleNet, and MobileNet. The results showed that L-CSMS achieved a competitive advantage with fewer parameters, FLOPs, and comparatively good accuracy.
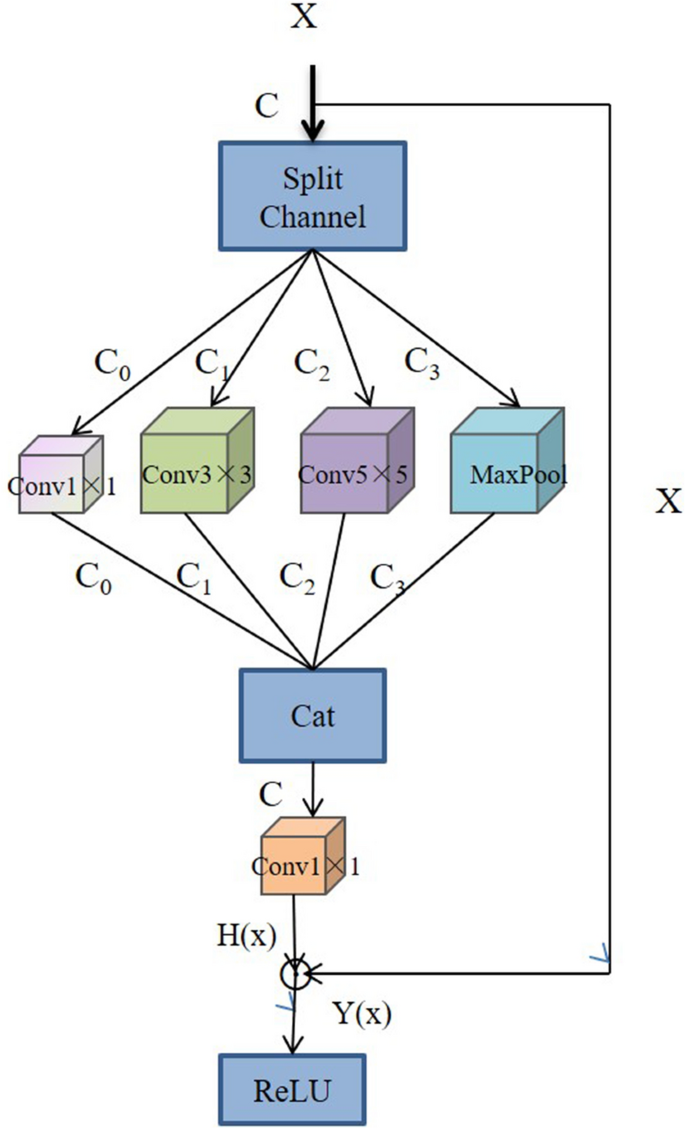
The building block with channel shuffle operation and multi-size convolution module 67 .
The ways to improve the CNN architecture are different, but the improvements are basically aimed at the same goal, which is to design a more accurate and practical plant disease severity assessment system with better generalization performance. Both studies focused on the residual structure in the ResNet and the channel shuffle.
CNN-based semantic segmentation network
Image semantic segmentation has received increasing attention from computer vision and DL researchers, and research work on semantic segmentation using DL techniques continues to evolve. In particular, CNN has far outperformed other methods in terms of accuracy and efficiency 69 . CNN-based segmentation provides not only category information, but also additional information about the spatial location of those categories. The task of semantic segmentation is to label each pixel as a kind of closed objects or a category of regions 70 . CNN-based segmentation theory has been applied to plant disease severity estimation and other related research in agriculture. The main goal of semantic segmentation applied to plant disease severity estimation is to assign appropriate labels to each pixel in order to obtain the percentage of diseased areas required for disease severity estimation.
Typically, the architecture of semantic segmentation is divided into two parts: the encoder network and the decoder network. The encoder is typically based on CNN networks to generate low-resolution image representations or feature maps that are mapped to pixel-level images and then perform prediction and segmentation. The differences between the different semantic segmentation models are often reflected in the decoder networks 70 . The first successful application of deep learning to semantic segmentation was achieved by a fully convolutional network (FCN) constructed by Long et al. 38 . After that, a number of variants of semantic segmentation emerged, such as U-Net, SegNet, DeepLab, and so on.
4 of the 16 articles used the CNN-based semantic segmentation network for plant disease severity assessment. Chen et al. 71 proposed a BLSNet for estimating the severity of rice bacterial leaf streak (BLS). BLSNet was based on U-Net with the addition of an attention mechanism and multi-scale extraction to improve the accuracy of lesion segmentation. Compared with DeepLabv3+ and U-Net, the experimental results suggested that BLSNet was more suitable for adapting to scale changes of images, and the prediction time of BLSNet was slightly longer than U-Net, but shorter than DeepLabV3+ . Gao et al. 72 proposed a SegNet-based network to segment potato late blight (PLB) lesions for quantification of the PLB severity. Goncalves et al. 73 conducted comparative experiments on six semantic segmentation networks (U-net, SegNet, PSPNet, FPN, and 2 variants of DeepLabv3+) applied to three types of plant disease severity estimation (coffee leaf miner, soybean rust, and wheat tan spot).
Although CNNs have provided good results in assessing the severity of plant diseases, the CNN-based semantic segmentation network also has its advantages. The achievement of CNN models for plant disease severity assessment is to directly establish a relationship between severity and samples that is applicable to certain plant diseases, but may not be appropriate for others. For other diseases, the model needs to be retrained. The CNN-based semantic segmentation network is a good solution to this problem by obtaining the percentage of diseased leaf area to reflect the severity through pixel-level segmentation. Previous studies have demonstrated the feasibility of a CNN-based semantic segmentation network for plant disease severity assessment.
The research from 2022
In the research of 2022, the methods of using CNN to evaluate the severity of plant diseases can be roughly divided into two categories. One is based on segmentation, and the other is based on improving the CNN, specifically adding the Attention Mechanism. In the segmentation evaluation method, the commonly used segmentation networks include DeepLabV3+ , U-Net, PSPNet and Mask R-CNN. For example, Zhang et al. 74 used the three-stage method to classify “Huangguan” pears. In the first stage, Mask R-CNN was used to segment “Huangguan” pears from complex backgrounds, and in the second stage, DeepLabV3+ , U-Net and PSPNet were used to segment the “Huangguan” pear spot, and the ratio of the spot area to the pixel area of the “Huangguan” pear was calculated, which was divided into three levels. In the third stage, ResNet-50, VGG-16 and MobileNetV3 were used to obtain the grade of “Huangguan” pear. Liu et al. 75 also used the idea of stage segmentation. Apple leaves were first segmented from the complex background using the deep learning algorithm, then the disease area of the segmented leaves was identified, and the ratio of the disease area to the leaf area was calculated to evaluate the severity of the disease. Instance segmentation can effectively separate the target from the complex background, which is conducive to dealing with the real environment. In the other method, Attention Mechanism has attracted people's attention. Yin et al. 76 improved the DCNN based on the addition of multi-scale and attention mechanism, and realized the classification of maize small leaf spot. Liu et al. 77 introduced a multi-scale convolution kernel and coordinate attention mechanism in SqueezeNext 78 to estimate disease severity, which was 3.02% higher than the original SqueezeNext model.
Datasets and evaluation metrics
Plant disease severity datasets.
The correct construction and rational use of plant disease severity datasets is a prerequisite and basis for severity assessment work. Unlike ImageNet, PlantVillage, and COCO in computer vision, there are no large unified datasets for plant disease severity. Plant disease severity datasets can be collected by taking one's own photographs and annotating the images, or by using public datasets and then annotating the images and citing other people's annotated images. With the development and popularity of electronic devices, image collection is typically done through cameras and smartphones. PlantVillage is a common public dataset used in plant disease severity, and common image annotation software is LabelMe, LabelImg, etc. This section provides links to datasets and annotation software used in the 16 studies, as shown in Table 1 .
Evaluation metrics
The common evaluation metrics mentioned in the previous model performance evaluation include accuracy, precision, recall, mean average precision (mAP), and F1 score based on precision and recall. Their specific definitions of them are described separately below.
Accuracy, Precision and Recall are expressed by the following equations:
In Eqs. ( 1 ) and ( 2 ), the true positive (TP), with a predicted value of 1 and an actual value of 1, indicates the number of correctly identified lesions. The false positive (FP), with a predicted value of 1 and an actual value of 0, indicates the number of misidentified lesions. The false negative (FN), with a predicted value of 0 and an actual value of 1, indicates the number of lesions not identified. The true negative (TN), with a predicted value of 0 and an actual value of 0, indicates the number of correctly identified non-lesions.
First, it is necessary to calculate the average precision for each category in the data set for mAP.
In the equation above, N is the number of all classes and j is the specific class in the dataset.
The average precision for each category is defined as follows:
The F1 score takes into account both the accuracy and recall of the model, and the equation is:
Challenges and future outlook
One of the most important and time-consuming parts of traditional machine learning (ML) methods is the manual feature extraction, while CNN can learn features automatically. Hedjazi MA et al. 86 addressed the task of visual identification of leaves in images by pre-training a CNN model. The experimental results showed that the pre-trained CNN model outperformed the classical machine learning methods using local binary patterns (LBPs). Bhujel A et al. 87 designed and tested a semantic segmentation model based on deep learning to detect and measure gray mold on strawberry plants. The results showed that the Unet model outperformed the traditional XGBoost, K-means, and image processing technologies in detecting and quantifying gray mold. Compared with traditional image processing methods and machine learning, plant disease severity assessment has broad application prospects and great development potential, either through the classical CNN framework, improved CNN architecture, or CNN-based semantic segmentation network. Although the technology of plant disease severity assessment is developing rapidly and has gradually moved from academic research to agricultural applications, there is still a certain gap from mature applications in real natural environments, and many problems need to be solved.
Dataset issues
Dataset problems can be divided into two main aspects: dataset insufficiency and dataset imbalance.
Dataset Insufficiency. Adequate datasets are necessary and fundamental for training the network. However, collecting and constructing datasets is an extremely time-consuming, labor-intensive, and costly process. Although there are a number of publicly available datasets for plant diseases, such as PlantVillage, ImageNet, and some publicly available self-made datasets. However, severity annotated datasets are really needed for plant disease severity research. Severity annotation of images is a more tedious process. There are two problems to face in the annotation process, one is the efficiency problem and the other is the accuracy problem. To address the time-consuming and complex annotation process that occurs in manual annotation, a possible solution is to automate the annotation with advanced software, and this automated annotation algorithm is urgently needed. In addition, semi-supervised training and auxiliary labeling methods can be used to increase the speed of agricultural sample processing and help alleviate the workload problem of manual semantic labeling. For the accuracy problem, errors are inevitable whether the annotation is done by manual visual assessment or by software, which is a challenge for future research 1 .
Dataset imbalance. The imbalance problem can have a serious impact on the performance of the model, for example, the misclassification rate becomes higher which has been demonstrated in the experiments of many studies 45 , 46 , 71 . This problem can be well mitigated by data augmentation and weighted loss functions 81 .
Complex background issues
The dataset can be divided into two types based on image backgrounds: images with uniform backgrounds taken under controlled conditions and images with complex backgrounds taken in natural environments. CNN models are more generalized by using images taken in a natural environment for training compared to a uniform background 56 , 57 , 58 . At the same time, complex backgrounds in images can cause other negative problems. For example, in realistic environments, ground stains resemble disease symptoms, leading to classification errors in the model 72 . Reflections from natural lighting can lead to misclassification of shaded healthy areas or failure to detect disease areas 79 . In addition, it is more common for multiple diseases to occur simultaneously in real-world environments. In 46 , the researchers mentioned that the presence of multiple diseases on a single sheet leaf can significantly change the characteristics of the symptoms, especially when the symptoms overlap, making the system more prone to misclassification. Many studies have shown that when disease symptoms are similar, their error separation rate increases significantly 45 , 51 , 60 . Due to the problems caused by the complex background, the application of the theoretical results of CNN-based plant disease severity assessment to the actual agricultural production process faces serious obstacles. To solve some of the problems caused by the complex environment, the images can be pre-processed, but this increases the complexity of the whole detection process. For the problem of simultaneous identification and assessment of multiple diseases, researchers in 46 proposed to alleviate this problem by training a similarity-based architecture that classifies symptoms that are not similar to any disease in the dataset into new classes, such as other classes. This idea has not yet been realized, and further solutions need to be brainstormed.
Practicality issues
In order to apply theoretical research to practical situations, various solutions have been proposed. As we all know, DCNN is an effective autonomous feature extraction model. Some researches combine deep learning and machine learning methods to build hybrid models. Usually, CNN is used as the feature extraction part and machine learning method is used as the classifier. Saberi Anari et al. 88 used improved CNN for feature extraction. And multiple Support Vector Machine (SVM) model was used to improve the speed of feature recognition and processing. Kaur et al. 89 used EfficientNet- B7 for feature extraction. After migration learning, they used logical regression technique to sample the collected features. Finally, the proposed variance technique was used to remove irrelevant features from the feature extraction vector. And the classification algorithm was used to classify the resulting features, and the most discriminative features are identified with the highest constant accuracy of 98.7%. By eliminating irrelevant features, the parameters of the model are greatly reduced. Vasanthan et al. 89 adopted AlexNet and VGG-19 for feature extraction, and selected the best subset of features by correlation coefficient, and fed them to K-nearest neighbor, SVM, Pulse Neutron Neutron (PNN), Fuzzy Logic, Artificial Neural Network (ANN) and other classifiers. The experimental results showed that the average accuracy of this method was more than 96%.
A server with supercomputing power is needed to ensure that the plant disease severity model built by CNN in the lab is widely used. Cloud computing is essentially a shared pool of computing resources. Cloud computing gathers many computing resources and realizes automatic management through software. Not limited by time and space, anyone who uses the Internet can use the huge computing resources and data centers on the network 90 . PaaS cloud is a concrete implementation of cloud computing. PaaS providers provide many infrastructure and other IT services, and users can access them anywhere through web browsers. The ability to pay for use allows organizations to eliminate the capital expenditures traditionally used for local hardware and software. Lanjewar et al. 91 deployed the CNN model used to evaluate tea diseases in the PaaS cloud, and the smartphone can access the hyperlink of the deployed model. The image of the tea can be captured by the smartphone camera and uploaded to the cloud. The cloud system automatically predicts the disease and displays it on the mobile display. Lanjewar M G et al. 92 used the PaaS cloud platform to deploy the CNN model for Curcuma longa detection. While cloud computing brings convenience to us, it inherits the security problems shared by computers and the Internet. In particular, privacy issues, resource theft, attack, and computer viruses. These potential security problems are serious and deserve our attention.
To deploy CNN models on the cloud computing platform, the smaller the size of the model, the better. However, whether the smaller the model can achieve the same evaluation effect is a question worth discussing. Increasing the model size to a certain extent shows better feature extraction effect, such as the comparison between AlexNet and DCNN models such as VGG and GoogLeNet. However, as the model becomes deeper and larger, the degradation problem occurs. The residual structure of ResNet effectively mitigates this problem. More and more lightweight networks have been proposed. Their efficiency may not be the best, but it is worth trading a small amount of effectiveness for a large amount of efficiency. Liu et al. 77 improved SquezeNext and performed comparative experiments with ReseNet-50, Xception, and MobileNet-V2. The experimental results showed that the accuracy of the proposed method was slightly better than that of Xconcept, while the model size was only 2.83 MB, which was only 3.45% of Xconcept. Model structure is a key factor to balance model size and performance.
Although CNN has shown excellent performance and great potential in assessing the severity of plant diseases, CNN also has its own limitations, such as translation invariance, pooling layer leading to information loss, and inability to obtain global features well. As a possible contribution and future work, new techniques that have become quite popular recently, such as vision transformers. The main feature of vision transformers 93 , 94 is the self-attention mechanism, which can capture the global information well. As far as I know, no research has applied it to severity estimation.
At present, some problems still have not found appropriate solutions, which indicates that the current research on automatic assessment of plant disease severity is far from mature and perfect practical application, which requires more scholars to continue to struggle to study the unsolved problems. The review article of our group on “Recent Advances in Plant Disease Severity Assessment Using Convolutional Neural Networks” provides some references for related types of research work. And more importantly, it can provide new ideas for the subsequent research work.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Bock, C. H. et al. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 29 (2), 59–107 (2010).
Article Google Scholar
Mahlein, A. K. et al. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 128 , 21–30 (2013).
Article ADS Google Scholar
Liang, Q. et al. (PDSE)-S-2-Net: Computer-assisted plant disease diagnosis and severity estimation network. Comput. Electron. Agric. 157 , 518–529 (2019).
Liu, L. et al. PestNet: An end-to-end deep learning approach for large-scale multi-class pest detection and classification. IEEE Access 7 , 45301–45312 (2019).
Madden, L. V., Hughes, G., Van Den Bosch, F. The study of plant disease epidemics (2007).
Cooke, B. Disease assessment and yield loss. In The Epidemiology of Plant Diseases 43–80 (Springer, 2006).
Chapter Google Scholar
Chiang, K. S. et al. Effects of rater bias and assessment method on disease severity estimation with regard to hypothesis testing. Plant. Pathol. 65 (4), 523–535 (2016).
Xu, Y. et al. Inheritance of resistance to zucchini yellow mosaic virus and watermelon mosaic virus in watermelon. J. Hered. 95 (6), 498–502 (2004).
Article CAS PubMed Google Scholar
Horsfall, J. & Cowling, E. Pathometry: The Measurement of Plant Disease Vol. 2 (Academic Press, 1978).
Google Scholar
Barratt, R. & Horsfall, J. J. P. An improved grading system for measuring plant disease. Phytopathology 35 , 655 (1945).
Forbes, G. & Korva, J. J. P. P. The effect of using a Horsfall-Barratt scale on precision and accuracy of visual estimation of potato late blight severity in the field. Plant Pathol. 43 (4), 675–682 (1994).
Bock, C. H. et al. The Horsfall-Barratt scale and severity estimates of citrus canker. Eur. J. Plant Pathol. 125 (1), 23–38 (2009).
Article MathSciNet Google Scholar
Bock, C. H. et al. From visual estimates to fully automated sensor-based measurements of plant disease severity: Status and challenges for improving accuracy. Phytopathol. Res. 2 (1), 1–30 (2020).
Del Ponte, E. M. et al. Standard area diagrams for aiding severity estimation: Scientometrics, pathosystems, and methodological trends in the last 25 years. Phytopathology 107 (10), 1161–1174 (2017).
Article PubMed Google Scholar
Cobb, N. A. Contributions to an economic knowledge of the Australian rusts (Uredineae). Agric. Gazette New South Wales 3 , 44–48 (1892).
McCulloch, W. S. & Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 5 (4), 115–133 (1943).
Article MathSciNet MATH Google Scholar
Fukushima, K. Neocognitron: A self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 36 (4), 193–202 (1980).
Article CAS MATH PubMed Google Scholar
Hubel, D. H. & Wiesel, T. N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 160 , 106–154 (1962).
Article CAS PubMed PubMed Central Google Scholar
Wang, W. et al. Development of convolutional neural network and its application in image classification: a survey. Opt. Eng. 58 (4), 040901–040901 (2019).
LeCun, Y. et al. Gradient-based learning applied to document recognition. Proc. IEEE 86 (11), 2278–2324 (1998).
Hinton, G. E., Osindero, S. & Teh, Y.-W. A fast learning algorithm for deep belief nets. Neural Comput. 18 (7), 1527–1554 (2006).
Article MathSciNet MATH PubMed Google Scholar
Glorot, X., Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics . JMLR Workshop and Conference Proceedings (2010).
Krizhevsky, A., Sutskever, I., Hinton, G. E. Imagenet classification with deep convolutional neural networks . Commun. ACM 25 (2012).
Simonyan, K., Zisserman, A.J. Very deep convolutional networks for large-scale image recognition (2014).
Szegedy, C., et al . Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2015).
He, K., et al . Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2016).
Huang, G., et al . Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017).
Iandola, F. N., et al . SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and< 0.5 MB model size (2016).
Howard, A.G., et al . Mobilenets: Efficient convolutional neural networks for mobile vision applications (2017).
Ma, N., et al . Shufflenet v2: Practical guidelines for efficient cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV) (2018).
Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017).
Tan, M., Lev Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In International Conference on Machine Learning (PMLR, 2019).
Girshick, R., et al . Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2014).
Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision (2015).
Ren, S., et al . Faster r-cnn: Towards real-time object detection with region proposal networks. 28 (2015).
Redmon, J., et al . You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2016).
Liu, W., et al . Ssd: Single shot multibox detector. In European Conference on Computer Vision (Springer, 2016).
Long, J., Shelhamer, E., Darrell, T. Fully convolutional networks for semantic segmentation . In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2015).
Badrinarayanan, V. et al. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39 (12), 2481–2495 (2017).
Ronneberger, O., Fischer, P., Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (Springer, 2015).
Zhao, H., et al . Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017).
Chen, L. -C., et al. , Semantic image segmentation with deep convolutional nets and fully connected crfs (2014).
He, K., et al . Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision (20170.
Wang, G. et al . Automatic image-based plant disease severity estimation using deep learning 2017 (2017).
Su, W. -H., et al . Automatic evaluation of wheat resistance to fusarium head blight using dual mask-RCNN deep learning frameworks in computer vision . Remote Sensing 13 (1) (2021).
Esgario, J. G. M., Krohling, R. A., Ventura, J. A. Deep learning for classification and severity estimation of coffee leaf biotic stress. Comput. Electron. Agric. 169 (2020).
Caruana, R. J. M. L. Multitask Learning , Vol. 28, 41–75 (1997).
Evgeniou, T., Pontil, M. Regularized multi-task learning. In Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (2004).
Ruder, S.J. An overview of multi-task learning in deep neural networks (2017).
Zhang, Y. & Yang, Q. A survey on multi-task learning. IEEE Trans. Knowl. Data Eng. 34 (12), 5586–5609 (2021).
Prabhakar, M. et al. Deep learning based assessment of disease severity for early blight in tomato crop. Multimed. Tools Appl. 79 (39), 28773–28784 (2020).
Zeng, Q. et al. Gans-based data augmentation for citrus disease severity detection using deep learning. IEEE Access 8 , 172882–172891 (2020).
Fenu, G. & Malloci, F. M. J. C. Using multioutput learning to diagnose plant disease and stress severity. Complexity 2021 , 1–11 (2021).
Mongeon, P. & Paul-Hus, A. J. S. The journal coverage of Web of Science and Scopus: A comparative analysis. Scientometrics 106 (1), 213–228 (2016).
Afzal, W., Torkar, R., Feldt, R. A systematic mapping study on non-functional search-based software testing. In Proceedings of the Twentieth International Conference on Software Engineering & Knowledge Engineering (SEKE'2008), San Francisco, CA, USA, July 1–3, 2008 (2008).
Goncharov, P. et al . Disease detection on the plant leaves by deep learning (2018).
Thapa, R., et al ., The plant pathology 2020 challenge dataset to classify foliar disease of apples (2020).
Zeng, W., et al . High-order residual convolutional neural network for robust crop disease recognition. In Proceedings of the 2nd International Conference on Computer Science and Application Engineering (2018).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
MATH Google Scholar
Mohanty, S. P., Hughes, D. P. & Salathé, M. J. F. Using deep learning for image-based plant disease detection. Front. Plant Sci. 7 , 1419 (2016).
Article PubMed PubMed Central Google Scholar
Ioffe, S., Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning . (PMLR, 2015).
Zeiler, M. D., Fergus, R. Visualizing and understanding convolutional networks . In European Conference on Computer Vision (Springer, 2014).
Chollet, F. Deep Learning with Python (Simon and Schuster, 2021).
Brahimi, M. et al. Deep learning for plant diseases: Detection and saliency map visualisation. In Human and Machine Learning 93–117 (Springer, 2018).
Sibiya, M. & Sumbwanyambe, M. J. P. Automatic fuzzy logic-based maize common rust disease severity predictions with thresholding and deep learning. Pathogens 10 (2), 131 (2021).
Boulch, A. J., Sharesnet: reducing residual network parameter number by sharing weights (2017).
Xiang, S. et al. L-CSMS: Novel lightweight network for plant disease severity recognition. J. Plant Dis. Protect. 128 (2), 557–569 (2021).
Szegedy, C., et al . Inception-v4, inception-resnet and the impact of residual connections on learning. In Thirty-First AAAI Conference on Artificial Intelligence (2017).
Garcia-Garcia, A., et al . A review on deep learning techniques applied to semantic segmentation (2017).
Hariharan, B., et al . Simultaneous detection and segmentation. In European Conference on Computer Vision (Springer, 2014).
Chen, S. et al. An approach for rice bacterial leaf streak disease segmentation and disease severity estimation. Agriculture 11 (5), 420 (2021).
Gao, J. et al. Automatic late blight lesion recognition and severity quantification based on field imagery of diverse potato genotypes by deep learning. Knowl.-Based Syst. 214 , 106723 (2021).
Goncalves, J. P. et al. Deep learning architectures for semantic segmentation and automatic estimation of severity of foliar symptoms caused by diseases or pests. Biosyst. Eng. 210 , 129–142 (2021).
Article CAS Google Scholar
Zhang, Y., et al . Appearance quality classification method of Huangguan pear under complex background based on instance segmentation and semantic segmentation . Front. Plant Sci. 13 (2022).
Liu, B.-Y. et al. Two-stage convolutional neural networks for diagnosing the severity of alternaria leaf blotch disease of the apple tree. Remote Sensing 14 (11), 2519 (2022).
Article ADS CAS Google Scholar
Yin, C. et al. Maize small leaf spot classification based on improved deep convolutional neural networks with a multi-scale attention mechanism. Agronomy 12 (4), 906 (2022).
Liu, Y., Gao, G. & Zhang, Z. J. I. A. Crop disease recognition based on modified light-weight CNN with attention mechanism. IEEE Access 10 , 112066–112075 (2022).
Gholami, A., et al . Squeezenext: Hardware-aware neural network design. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (2018).
Ozguven, M. M. Deep learning algorithms for automatic detection and classification of mildew disease in cucumber. Fresenius Environ Bull 29 (8), 7081–7087 (2020).
CAS Google Scholar
Hu, G. et al. Detection and severity analysis of tea leaf blight based on deep learning. Comput. Electric. Eng. 90 , 107023 (2021).
Fernández-Campos, M. et al. Wheat spike blast image classification using deep convolutional neural networks. Front. Plant Sci. 12 , 1054 (2021).
Hayit, T. et al. Determination of the severity level of yellow rust disease in wheat by using convolutional neural networks. J. Plant Pathol. 103 (3), 923–934 (2021).
Hughes, D., Salathé, M. J. An open access repository of images on plant health to enable the development of mobile disease diagnostics (2015).
Al Gubory, K. H. J. B. S. O. A. Plant polyphenols, prenatal development and health outcomes. Biol. Syst. Open Access 3 (1), 1–2 (2014).
Franceschi, V. T. et al. A new standard area diagram set for assessment of severity of soybean rust improves accuracy of estimates and optimizes resource use. Plant Pathol. 69 (3), 495–505 (2020).
Hedjazi, M. A., Kourbane, I., Genc, Y. On identifying leaves: A comparison of CNN with classical ML methods. In 2017 25th Signal Processing and Communications Applications Conference (SIU) (IEEE, 2017).
Bhujel, A. et al ., Detection of gray mold disease and its severity on strawberry using deep learning networks . J. Plant Dis. Protect. 1–14 (2022).
Saberi Anari, M. J A hybrid model for leaf diseases classification based on the modified deep transfer learning and ensemble approach for agricultural AIoT-Based monitoring. Comput. Intell. Neurosci. 2022 (2022).
Kaur, P. et al. Recognition of leaf disease using hybrid convolutional neural network by applying feature reduction. Sensors 22 (2), 575 (2022).
Article ADS MathSciNet PubMed PubMed Central Google Scholar
Huth, A., Cebula, J. The basics of cloud computing. U. S. Comput. 1–4 (2011).
Lanjewar, M. G., Panchbhai, K.G., Convolutional neural network based tea leaf disease prediction system on smart phone using paas cloud. Neural Comput. Appl. 1–17 (2022).
Lanjewar, M. G. et al. Detection of tartrazine colored rice flour adulteration in turmeric from multi-spectral images on smartphone using convolutional neural network deployed on PaaS cloud. Multimed. Tools Appl. 81 (12), 16537–16562 (2022).
Dosovitskiy, A., et al ., An image is worth 16x16 words: Transformers for image recognition at scale (2020).
Xie, E. et al. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 34 , 12077–12090 (2021).
Download references
Acknowledgements
We deeply acknowledge The National Natural Science Foundation of China, Natural Science Foundation of Hunan Province China, and Hunan Provincial Education Science “13th Five-Year Plan” Foundation.
This research is funded by The National Natural Science Foundation of China, No. 31870532, Natural Science Foundation of Hunan Province China, No. 2021JJ31163, Hunan Provincial Education Science “13th Five-Year Plan” Fund, No. XJK20BGD048.
Author information
Authors and affiliations.
College of Computer and Information Engineering, Central South University of Forestry and Technology, Changsha, 410004, China
Tingting Shi, Yongmin Liu, Kui Hu, Hao Huang, Hanlin Liu & Hongxu Huang
Research Center of Smart Forestry Cloud, Central South University of Forestry and Technology, Changsha, 410004, China
Business School of Hunan Normal University, Changsha, 410081, China
Xinying Zheng
You can also search for this author in PubMed Google Scholar
Contributions
T. S.: Be responsible for writing, revising papers, drawing charts, and revision of articles. Y. L.: put forward the main research problems of this paper, control the writing progress of the paper and participate in the revision of the paper. X. Z.: reviewed the manuscript. K. H.: reviewed the manuscript. Hao Huang: reviewed the manuscript. H. L.: reviewed the manuscript. Hongxu Huang: reviewed the manuscript.
Corresponding author
Correspondence to Yongmin Liu .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Shi, T., Liu, Y., Zheng, X. et al. Recent advances in plant disease severity assessment using convolutional neural networks. Sci Rep 13 , 2336 (2023). https://doi.org/10.1038/s41598-023-29230-7
Download citation
Received : 28 July 2022
Accepted : 31 January 2023
Published : 09 February 2023
DOI : https://doi.org/10.1038/s41598-023-29230-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Revolutionizing crop disease detection with computational deep learning: a comprehensive review.
- Habiba N. Ngugi
- Absalom E. Ezugwu
- Laith Abualigah
Environmental Monitoring and Assessment (2024)
Classification of Plant Leaf Disease Using Deep Learning
Journal of The Institution of Engineers (India): Series B (2024)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.
REVIEW article
An advanced deep learning models-based plant disease detection: a review of recent research.
A correction has been applied to this article in:
Corrigendum: An advanced deep learning models-based plant disease detection: a review of recent research
- Read correction

- 1 Department of Computer Science, CECOS University of IT and Emerging Sciences, Peshawar, Pakistan
- 2 Department of Computer Science and Information Technology, Sarhad University of Science and Information Technology, Peshawar, Pakistan
- 3 College of Technological Innovation, Zayed University, Dubai, United Arab Emirates
- 4 Faculty of Computer Science and Engineering, Galala University, Suez, Egypt
- 5 Information Systems Department, Faculty of Computers and Artificial Intelligence, Benha University, Banha, Egypt
- 6 Department of Molecular Stress Physiology, Center of Plant Systems Biology and Biotechnology, Plovdiv, Bulgaria
- 7 Department of Electrical Engineering, College of Engineering, Jouf University, Jouf, Saudi Arabia
- 8 Department of Plant Physiology and Molecular Biology, University of Plovdiv, Plovdiv, Bulgaria
- 9 School of Computer Science and Information Engineering, Zhejiang Gongshang University, Hangzhou, China
- 10 Department of Computer Science and Engineering, School of Convergence, College of Computing and Informatics, Sungkyunkwan University, Seoul, Republic of Korea
Plants play a crucial role in supplying food globally. Various environmental factors lead to plant diseases which results in significant production losses. However, manual detection of plant diseases is a time-consuming and error-prone process. It can be an unreliable method of identifying and preventing the spread of plant diseases. Adopting advanced technologies such as Machine Learning (ML) and Deep Learning (DL) can help to overcome these challenges by enabling early identification of plant diseases. In this paper, the recent advancements in the use of ML and DL techniques for the identification of plant diseases are explored. The research focuses on publications between 2015 and 2022, and the experiments discussed in this study demonstrate the effectiveness of using these techniques in improving the accuracy and efficiency of plant disease detection. This study also addresses the challenges and limitations associated with using ML and DL for plant disease identification, such as issues with data availability, imaging quality, and the differentiation between healthy and diseased plants. The research provides valuable insights for plant disease detection researchers, practitioners, and industry professionals by offering solutions to these challenges and limitations, providing a comprehensive understanding of the current state of research in this field, highlighting the benefits and limitations of these methods, and proposing potential solutions to overcome the challenges of their implementation.
1 Introduction
The use of ML and DL in plant disease detection has gained popularity and shown promising results in accurately identifying plant diseases from digital images. Traditional ML techniques, such as feature extraction and classification, have been widely used in the field of plant disease detection. These methods extract features from images, such as color, texture, and shape, to train a classifier that can differentiate between healthy and diseased plants. These methods have been widely used for the detection of diseases such as leaf blotch, powdery mildew, and rust, as well as disease symptoms from abiotic stresses such as drought and nutrient deficiency ( Mohanty et al., 2016 ; Anjna et al., 2020 ; Genaev et al., 2021 ) but have limitations in accurately identifying subtle symptoms of diseases and early-stage disease detection. In addition, they also struggle to process complex and high-resolution images.
Recently, DL techniques such as convolutional neural networks (CNNs) and deep belief networks (DBNs) have been proposed for plant disease detection ( Liu et al., 2017 ; Karthik et al.,2020 ). These methods involve training a network to learn the underlying features of the images, enabling the identification of subtle symptoms of diseases that traditional image processing methods may not be able to detect ( Singh and Misra, 2017 ; Khan et al., 2021 ; Liu and Wang, 2021b ). DL models can handle complex and large images, making them suitable for high-resolution images ( Ullah et al., 2019 ). However, these methods require a large amount of labeled training data and may not be suitable for unseen diseases. Furthermore, DL models are computationally expensive, which may be a limitation for some applications.
In recent years, several research studies have proposed different ML and DL approaches for plant disease detection. However, most studies have focused on a specific type of disease or a specific plant species. Therefore, more research is needed to develop a generalizable and robust model that can work for different plant species and diseases. Additionally, there is a need for more publicly available datasets for training and evaluating models. One of the recent trends in the field is transfer learning, a technique that allows for reusing pre-trained models on new datasets. Recently, transfer learning and ensemble methods have emerged as popular trends in plant disease detection using ML and DL. Transfer learning involves fine-tuning pre-trained models on a specific dataset to enhance the performance of DL models. Ensemble methods, on the other hand, involve combining multiple models to improve overall performance and reduce dependence on a single model. These approaches have been applied to increase the robustness and accuracy of plant disease detection models. Additionally, it can also prevent overfitting, a common problem in DL models where the model performs well on the training data but poorly on unseen data. Another essential aspect to consider is the use of data augmentation techniques, which is the process of artificially enlarging the size of a dataset by applying random transformations to the images. This approach has been used to increase the diversity of the data and reduce the dependence on a large amount of labeled data.
In conclusion, the application of ML and DL techniques in plant disease detection is a rapidly evolving field with promising results. While these techniques have demonstrated their potential to accurately identify and classify plant diseases. There are still limitations and challenges that need to be addressed. Further research is required to develop generalizable models and make more publicly available datasets for training and evaluation. This review highlights the current state of research in this field and provides a comprehensive understanding of the benefits and limitations of ML and DL techniques for plant disease detection. Its novelty lies in the breadth of coverage of research published from 2015 to 2022, which explores various ML and DL techniques while discussing their advantages, limitations, and potential solutions to overcome implementation challenges. By offering valuable insights into the current state of research in this area, the article is a valuable resource for plant disease detection researchers, practitioners, and industry professionals seeking a thorough understanding of the subject matter.
The following section comprises the contributions of this research article.
● This paper provides an overview of the current developments in the field of plant disease detection using ML and DL techniques. By covering research published between 2015 and 2022, it provides a comprehensive understanding of the state-of-the-art techniques and methodologies used in this field.
● This review examines various ML and DL methods for detecting plant diseases, including image processing, feature extraction, CNNs, and DBNs, and sheds light on the benefits and drawbacks, such as data availability, imaging quality, and differentiation between healthy and diseased plants. The article shows that the use of ML and DL techniques significantly increases the precision and speed of plant disease detection.
● Various datasets related to plant disease detection have been studied in the literature, including PlantVillage, the rice leaf disease dataset, and datasets for insects affecting rice, corn, and soybeans.
● The paper discussed various performance evaluation criteria used to assess the accuracy of plant disease detection models, including the intersection of unions (IoU), dice similarity coefficient (DSC), and accurate recall curves.
The article has seven main sections. A brief overview of plant disease and pest detection and its significance is provided in Section 1. The challenges and issues in the plant disease and pest detection are discussed in Section 2. The deep learning approaches for recognizing images and their applications in plant disease and pest detection are presented in Section 3. The comparison of commonly used datasets and the performance metrics of deep learning methods on different datasets are presented in Section 4. The challenges in existing systems are identified in Section 5. The discussion about the identification of plant diseases and pests is presented in Section 6. Finally, the conclusion of the research work and future research directions are discussed in Section 7.
2 Plant disease and pest detection: Challenges and issues
2.1 identifying plant abnormalities and infestations.
Artificial Intelligence (AI) technologies have recently been applied to the field of plant pathology for identifying plant abnormalities and infestations. These technologies can have the capability to transform the method in which plant maladies are identified, diagnosed, and managed. In this passage, we will explore the various AI technologies that have been proposed for identifying plant abnormalities and infestations, their advantages and limitations, and the impact of these technologies on the field of plant pathology. One of the most widely used AI technologies in plant pathology is ML. ML algorithms, such as c4.5 classifier, tree bagger, and linear support vector machines, have been applied to the classification of plant diseases from digital images. These algorithms can be trained to recognize specific patterns and symptoms of diseases, making them suitable for the classification of diseases in their primary phases. However, ML algorithms mandate a substantial quantity of data that has been annotated for training and may not be suitable for diseases that have not been seen before.
DL technologies, such as CNNs and DBNs, have also been proposed for identifying plant abnormalities and infestations. These technologies have been showing promising outcomes in the detection and identification of lesions from digital images ( Kaur and Sharma, 2021 ; Siddiqua et al., 2022 ; Wang, 2022 ). DL models can automatically learn features from the images and can identify subtle symptoms of diseases that traditional image processing methods may not be able to detect. Though, Deep Learning models necessitate a significant volume of labeled training data and involve intensive computational resources, which may be a limitation for some applications. Another AI technology that has been applied to plant pathology is computer vision (CV). CV algorithms, such as object detection and semantic segmentation, can be used to identify and localize specific regions of interest in images, such as plant leaves and symptoms of diseases ( Kurmi and Gangwar, 2022 ; Peng and Wang, 2022 ). These algorithms can be used to automatically transforming the images into recognizable patterns or characteristics can be integrated with ML or DL algorithms for disease detection and classification. However, CV algorithms need a huge number of labeled image data for model training and may not be suitable for diseases that have not been seen before. Figure 1 comprises four images, each depicting a different stage of plant disease detection. The first image is the input image, while the next image displays the disease identification results. The third image features lesion detection, and the final image presents the segmentation results of the plant lesion.
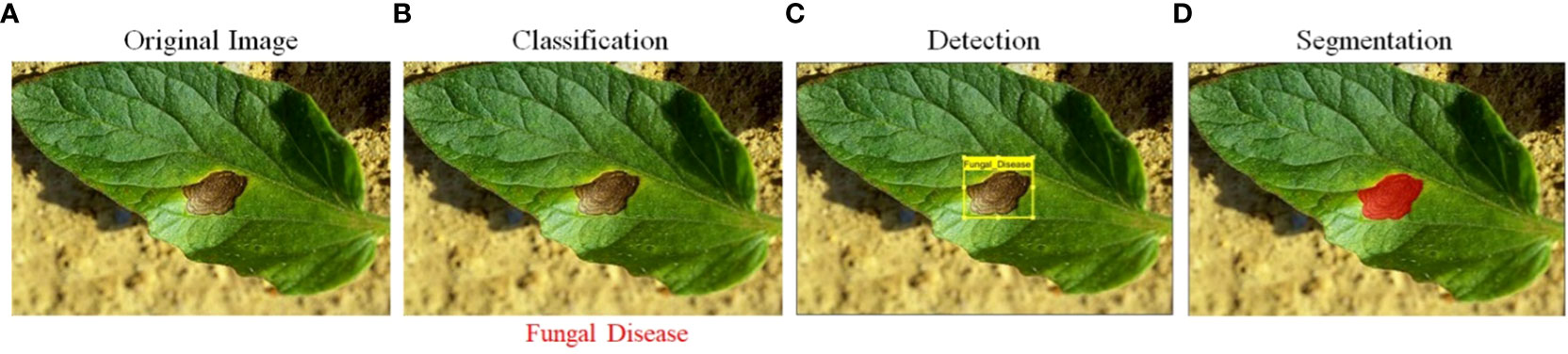
Figure 1 (A) Input raw image, (B) leaf classification, (C) lesion detection, and (D) lesion segmentation.
AI technologies have shown promising results in identifying plant abnormalities and infestations. ML, DL, and CV based system are utilized for to the classification and lesion segmentation of plant diseases from digital images and could change the method of discovering plant illnesses significantly, diagnosed, and managed ( Akbar et al., 2022 ). However, these technologies need a considerable amount of annotated training data and may not be suitable for diseases that have not been seen before. Further research is needed to develop generalizable models that can be applied to different plant species and diseases, and to make more datasets publicly available for training and evaluating the models. Table 1 provides comprehensive information about the tools and technologies utilized for plant disease detection. It includes details about the various feature extraction methods, including those based on handcrafted and learning features, as well as the appropriate methods for processing small and large plant image datasets.
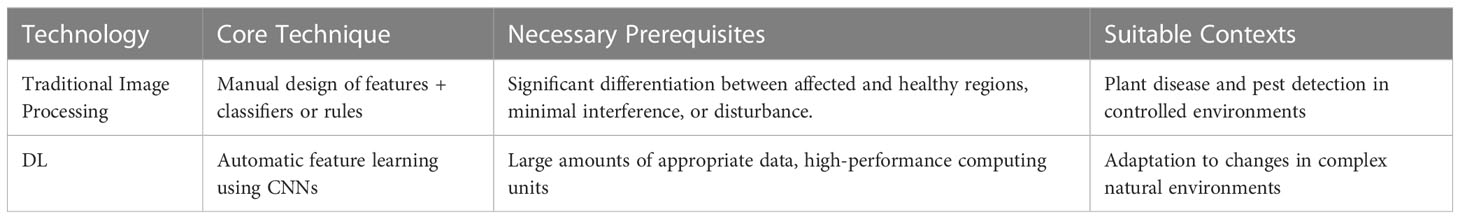
Table 1 Comparison of different technologies for image processing.
2.2 Evaluation of conventional techniques for identifying plant diseases and pests
In recent years, ML and DL-based approaches have been increasingly applied to agriculture and botanical studies. These approaches have shown great potential in improving crop yield, identifying plant lesions, and optimizing plant growth. In comparison to traditional approaches, ML and DL-based methods offer several advantages and have the potential to revolutionize the field of agriculture and botanical studies. Traditional approaches in agriculture and botanical studies mainly rely on manual inspection and expert knowledge. These methods are often time-consuming, physically demanding, and susceptible to human mistakes. In contrast, ML and DL-based approaches can automate these tasks, reducing the need for human interference and enhancing precision and efficiency of the process.
ML and DL-based approaches have been used to analyze large amounts of data, including images, sensor data, and weather data, to identify patterns and make predictions. For example, ML algorithms such as c4.5 classifier and tree bagger are being used to predict crop yields, identify plant lesions and pests, and optimize plant growth ( Yoosefzadeh-Najafabadi et al., 2021 ; Cedric et al., 2022 ; Domingues et al., 2022 ). DL models, such as CNNs and DBNs, have been applied plant lesion identification based on image analysis and classification, providing better accuracy and robustness compared to traditional image processing methods ( Sladojevic et al., 2016 ; Alzubaidi et al., 2021 ; Dhaka et al., 2021 ). The ML and DL-based approaches offer several advantages over traditional methods in agriculture and botanical studies. These methods can automate tasks, increase accuracy and efficiency, and analyze huge quantity of data. Since, these methods require a large size of labeled features and may not be suitable for lesions that have not been seen before. Further research is needed to develop generalizable models that can be applied to different crop species and conditions, and to make more datasets publicly available for predictive model training and model validation for performance analysis.
3 Deep learning approaches for recognizing images
DL approaches have become a promising method for detecting plant lesions. These techniques, which are based on RNN have demonstrated success by achieving high accuracy in identifying various plant lesions from images ( Xu et al., 2021 ). By automatically learning features from the images, DL models can accurately identify and classify different disease symptoms, reducing the need for manual feature engineering ( Drenkow et al., 2021 ). Additionally, these models can handle large amounts of data, making them well-suited for large-scale plant lesions detection ( Arcaini et al., 2020 ). Therefore, in review paper, we evaluate the current state-of-the-art in using DL for plant lesions recognition, examining various architectures, techniques, and datasets used in this field. Our aim is to provide a thorough understanding of the current research in this area and identify potential future directions for improving the detection precision and make the identification system more efficient using the DL approaches.
3.1 Deep learning theory
Iqbal ( Sarker, 2021 ) popularized the term “Deep Learning” in a 2006 Science article (DL). The article describes a procedure for transforming high-dimensional data into low-dimensional codes using a technique called “autoencoder” networks. These networks are made up of a layers with few parameters that is trained to create vectors of input with high dimensions. The process of fine-tuning the weights of the network can be done using gradient descent, but this method is only effective if the baseline weights are near to a satisfactory solution. The article presents an effective initialization of weights that enables deep autoencoder models to learn the low-dimensional sequences that are more effective than principal component analysis for reducing the dimensionality of data.
DL is a variant of ML that employs multiple-layered AI networks to learn and represent complex patterns in data. It is extensively employed in object recognition, object detection, speech analysis and speech-to-text transcription. In natural language processing, DL-based models are used for tasks such as language translation, text summarization, and sentiment analysis. Additionally, DL is also used in recommendation systems to predict user preferences based on previous actions or interactions. AI vision is a subfield of artificial intelligence concerned with the construction of computers to process and understand the visual contents from the world ( Liu et al., 2017 ).
In traditional manual image classification and recognition methods, the underlying characteristics of an image are extracted through the use of hand-crafted features. These methods, however, are limited in their ability to extract information about the deep and complex characteristics of an image. This is because the manual extraction procedure is extremely reliant on the expertise of an individual conducting the analysis, and can be prone to errors and inconsistencies. Additionally, traditional manual methods are not able to extract information about subtle or hidden features that may be present in an image. In contrast, DL-based image classification and recognition methods use artificial neural networks to automatically extract image features. These methods have been shown to be highly effective in extracting complex and deep features from images, and have been utilized in numerous applications such as object recognition, facial features recognition, and image segmentation. Among the primary benefits of DL-based methods is its capacity to learn features autonomously from input data, rather than relying on manual feature engineering. This allows the model to learn more abstract and subtle features that may be present in the image, leading to improved performance and greater accuracy. Additionally, DL-based methods are also able to handle high-dimensional and complex data, making them particularly well-suited to handling large-scale image datasets. In summary, traditional manual image classification and recognition methods have limitations in extracting deep and complex characteristics of an image, while DL-based methods have been demonstrated greater efficiency and effectiveness in this task by automatically extracting image features, handling high-dimensional and complex data, and learning more abstract and subtle features that may be present in the image ( Tran et al., 2015 ).
DBN ( Hasan et al., 2020 ) is a type of unsupervised DL model that is composed of multiple layers of Restricted Boltzmann Machines (RBMs). Using the plant lesion and pest infestation detection, DBNs have been used to test plant images affected regions to detect various diseases and types of pests, and extract features from images of plant leaves. Studies have shown that DBNs can achieve high accuracy rates in the range of 96-97.5% in classifying images of plant leaves affected by diseases and pests.
Boltzmann’s Deep Machine (DBM) ( Salakhutdinov & Larochelle, 2010 ) is generative stochastic AI model that can be utilized for unsupervised classification to detect the plant lesion. Within the context of conventional plant lesion and pest detection, DBMs have been used to predict labels for images of various plant affected regions by viruses and plant bugs, and extract features from images of plant leaves. Studies have shown that DBMs can achieve high accuracy rates in the range of 96-96.8% in classifying images of plant leaves affected by diseases and pests.
Deep Denoising Autoencoder ( Lee et al., 2021 ) is a variant of autoencoder, which is a neural network architecture that is composed of an encoder module along with a decoder. In the context of traditional plant disease and pest infestation detection, DDA has been used to for two different purposed i.e., noise removal from the plant leaf data and a prediction system to identify plant disease. Studies have shown that DDA can achieve high accuracy rates in the range of 98.3% in classifying images of plant leaves affected by diseases and pests.
Deep CNN ( Shoaib et al., 2022a ; Shoaib et al., 2022b )is a type of feedforward AI model that is consisting of several hidden layers of convolutional and pooling layers, the CNN model are the best of the DL model for achieving higher detection accuracy using imaging data The CNN model consist of two blocks, the features learning and classification blocks. The features learning block extract various kind of features using the convolutional layer where the features learning is performed at the fully connected layers. The higher accuracy of the CNN model for plant disease classification has proofed to be the best then all other kinds of ML and DL methods. Studies have shown that CNNs can achieve high accuracy rates in the range of 99-99.2% in classifying images of plant leaves affected by diseases and pests.
3.2 Convolutional neural network
CNNs are a sort of DL model that are ideally suited for image classification tasks such as leaf disease detection ( Zhang et al., 2019 ; Lin et al., 2020 ; Stančić et al., 2022 ). Multiple layers comprise the CNN’s architecture, such as fully connected layers, maxpooling, and normalization layers. The first layer in the CNN is the input layer while the second layer in most of the CNNs is convolutional layers which extract features by applying various kind of 2D filters on the image, the amount of images increase which can then dimensionally reduced pooling also known as down sampling layers, resulting in a more compact representation of the image. Fully connected (FC) layers in a CNN are also known as learnable features, the extracted features are processed in the FC layer for learning and weights optimization. These layers are also responsible for making classification which can be used to recognize various plant diseases. The learning process of CNN model begins with training, the input to the CNN are images along with their labels, after the successful training of the model, the model is able to identify disease types.
The decision-making process in a CNN for leaf disease detection starts with the input of an image of a leaf. The image is then passed through the convolutional layers, where features are extracted. The feature vectors are then processed by pooling layers, where the spatial dimensions are reduced. The feature vectors are then transmitted via the FC layers, where a decision is made about the presence of a disease or pest. The models output are the probabilities that the leaf is diseased or healthy. CNNs are well-suited for leaf disease detection, thanks to their architecture consisting of up-sampling, down-sampling and learnable layers ( Agarwal et al., 2020 ). The learning process of CNN involves training the network using labeled images of healthy and disease effected plants. Figure 2 presents a framework for classifying the plants into normal and abnormal plant using leaf data. The framework employs several different Inception architectures, and the final decision is made through a bagging-based approach.
Figure 2 A CNN framework for classifying plants into healthy and unhealthy ( Shoaib et al., 2022a ).
3.3 Deep learning using open-source platforms
TensorFlow is a powerful library for dataflow and differentiable programming ( Abadi, 2016 ; Dillon et al., 2017 ), which allows for efficient computation on a set of devices with powerful hardware’s, that include memory, GPUs and TPUs. Its ability to create dataflow graphs, which describe how data moves through a computation, makes it a popular choice for ML and DL applications. In contrast, Keras is a high-end DL library that operates atop TensorFlow (also some other libraries). It simplifies the creation of DL models by providing a user-friendly API, and it provides a number of pre-built layers and functions, such as convolutional layers and pooling layers, which can be easily added to a model. In recent versions of Tensorflow (2.4 and above). TensorFlow is used to provide low-level operations for building and training models, while Keras is used to provide a higher-level API for building and training models more easily. The use of TensorFlow and Keras together in this research has allowed us to effectively and efficiently solve the problem at hand.
PyTorch is also from the open-source community which has lot of capabilities for developing ML and DL applications ( Zhao et al., 2021 ; Masilamani and Valli, 2021 ). PyTorch is a powerful library for building and training DL models. It is known for its flexibility and ease of use, making it a popular choice among researchers and practitioners. One of the key features of PyTorch is its dynamic computational graph. Unlike other libraries, such as TensorFlow, which uses a static computational graph, PyTorch allows for the modification of the graph on-the-fly, making it more suitable for research and experimentation. Additionally, PyTorch provides support for distributed training, allowing for efficient training of large models on multiple GPUs. PyTorch also provides a number of pre-built modules, such as convolutional layers and recurrent layers, which can be easily added to a model. This makes it easy to quickly prototype and experiment with different model architectures. Additionally, PyTorch also has a large community that shares pre-trained models, datasets, and tutorials, which helps to make the development process even more efficient.
Caffe (Convolutional Architecture for Fast Feature Embedding) is a Berkeley Vision and Learning Center-developed open-source DL framework (BVLC) and community contributors ( Jia et al., 2014 ). It is a popular choice for image and video classification tasks such as object detection and video summarization, and also consider a good choice for its speed and efficiency in training large models. Caffe is implemented in C++ and has a Python interface, which allows for easy integration with other Python libraries such as NumPy and SciPy. This allows for a high level of flexibility in the design and experimentation of DL models. One of the key features of Caffe is its ability to perform efficient convolutional operations, which are essential for computer vision tasks. Additionally, Caffe supports a wide range of DL models, such as CNN, RNN, Transformers networks. It also provides a number of pre-built layers and functions, such as convolutional layers and pooling layers, which can be easily added to a model ( Komar et al., 2018 ).
The Montreal Institute for Learning Algorithms (MILA) at the University of Montreal created Theano which also covers the open source license and have several packages in the python language for ML and DL ( Bahrampour et al., 2015 ). It is widely used for DL and other numerical computations, and it is known for its ability to optimize and speed up computations on CPUs and GPUs. One of the key features of Theano is its ability to perform symbolic differentiation, which allows for the efficient computation of gradients during the training of DL models ( Chung et al., 2017 ). Additionally, Theano can automatically optimize computations and perform automatic differentiation, which allows for the efficient training of large models. Theano also provides a number of pre-built functions, such as convolutional and recurrent layers, which can be easily added to a model. Theano is implemented in Python, which allows for easy integration with other Python libraries such as NumPy and SciPy. This allows for a high level of flexibility in the design and experimentation of DL models.
Table 2 in the research article provides a comparison of several popular Artificial Intelligence (AI) frameworks. The table compares the technology, developer, auxiliary devices required, functionality, programming language, and popular applications of each framework. This information is valuable for researchers and practitioners in the field of AI, as it provides an overview of the various options available and the strengths and limitations of each framework. The data presented in Table 2 can be used to guide the selection of an appropriate AI framework for a specific task or application.

Table 2 Comparison of popular artificial intelligence frameworks.
3.4 Deep learning based plant lesion and pests detection system
This section of the research focuses on the application of DL methods for segmentation plant lesions and pest infestation in botany and agriculture. With the increasing demand for food and the need for sustainable agricultural practices, the prompt identification and handling of illnesses affecting plants and pests is crucial for ensuring crop yields and maintaining the health of crops. DL, with its ability to process large amounts of data and its ability to learn from the data, has proven to be a robust tool for detecting plant diseases and pest infestation. In this section, we present a comprehensive overview of the state-of-the-art DL methods that have been developed for this purpose, including methods for image-based disease and pest detection, as well as methods for data-driven disease and pest detection using sensor data and other types of data. We also discuss the challenges and limitations of these methods and provide insights into future research directions. In particular, we will cover the recent advancements in DL for disease and pest detection, including the use of CNN, recurrent neural networks, and transfer learning techniques. These DL methods have shown to be effective in detecting plant diseases and pest infestation at a high level of accuracy, which can support farmers and agricultural professionals in taking appropriate action to prevent crop losses.
3.4.1 Classification network
Various Convolutional Neural Network (CNN) models which have been utilized to identify plant diseases and pest infestation are discussed. The first model that we will discuss is AlexNet ( Antonellis et al., 2015 ), which is the CNN model developed in 2012. The AlexNet CNN win the classification challenge by achieving the highest accuracy using the 1000 classes Imagenet dataset. AlexNet is known for its high accuracy and speed, and it has been used for a variety of tasks, including plant disease detection. Another popular CNN model is VGG ( Soliman et al., 2019 ), which was established in 2014 by the University of Oxford’s at Visual Geometry Lab. VGG is known for its high accuracy and is often used for image classification tasks. It has been employed to detect plant lesions by extracting hidden patterns from plant leaf data.
ResNet ( Szymak et al., 2020 ), which was developed by Microsoft Research Asia in 2015, is known for its ability to handle very deep networks. It has been used for plant disease detection by using pre-trained ResNet models on the images of the plants. GoogLeNet ( Wang et al., 2015 ), which was developed by Google in 2014, is known for its high accuracy and efficient use of computation resources. It has been used for plant disease detection by fine-tuning pre-trained GoogLeNet models on the images of the plants. InceptionV3, which was developed by Google in 2015, is known for its high accuracy and efficient use of computation resources. It has been used for plant disease detection by fine-tuning pre-trained InceptionV3 models on the images of the plants. DenseNet ( Tahir et al., 2022 ), which was developed in the ( Huang et al., 2017 ), is known for its ability to handle very deep networks and efficient use of computation resources. It has been used for plant disease detection by fine-tuning pre-trained DenseNet models on the images of the plants. These CNN models differ in their architectures, sizes, shapes, and the number of parameters. While AlexNet, VGG, GoogLeNet, InceptionV3, and DenseNet have been widely used for plant disease detection, ResNet is known for its ability to handle very deep networks. All these models have been shown to be effective in detecting plant diseases and pests based on different characteristics such as size, shape, and color, and they can be employed for harvesting characteristics from pictures of the plants which can be used to train a classifier to detect different diseases and pests.
3.4.2 CNN as features descriptor
The article ( Sabrol, 2015 ) “Recent Research on Image Processing and Soft Computing Approaches for Identifying and Categorizing Plant Diseases using CNNs” discusses the use of CNNs for recognizing and classifying plant diseases. The authors review various studies that have used CNNs, which are a type of DL algorithm, to detect and diagnose plant diseases. They also discuss the challenges and limitations of using CNNs, such as the need for large amounts of data, the high computational requirements, and the potential for overfitting. The article concludes by highlighting the potential for further research in this area and the importance of developing accurate and reliable plant disease recognition and classification systems using CNNs.
This research article presents an architecture of Convolutional Neural Networks for determining the variety of crops from image sequences obtained from advanced agro-observation stations ( Yalcin and Razavi, 2016 ). The authors address challenges related to lighting and image quality by implementing preprocessing steps. They then employ the CNN architecture to extract features from the images, highlighting the importance of the construction and depth of the CNN architecture in determining the recognition capability of the network. The accuracy of the model presented is evaluated to perform a comparison between the CNN model with those obtained using a support vector machine (SVM) classifier with the utilization of feature extractors such as Local Binary Patterns (LBP) and Gray-Level Co-Occurrence Matrix. The results of the approach are tested on a dataset collected through a government-supported project in Turkey, which includes over 1,200 agro-stations. The experimental outcomes affirm the efficiency of the suggested technique.
A novel meta-architecture is proposed, which utilizing a CNN designed for distinguishing between healthy and diseased plants ( Fuentes et al., 2017b ). The authors employed multiple characteristic extractors within the CNN to analyze input images that are divided into their corresponding categories. On the other hand, a CNN-based approach for the identification of various eight classes of rice viruses is presented in ( Hasan et al., 2019 ). The authors performed features extraction using the features learning model and introduced them along with the corresponding labels into a support vector machine (SVM) linear multiclass model for training. The trained model achieved a validation accuracy of 97.5%.
3.4.3 CNN-based predictive systems
In the area of plant illness and pest identification, CNNs have been extensively utilized. One of the first applications of CNNs in this field was the identification of lesions in plant images, utilizing classification networks. The method employed involves training CNNs to recognize specific patterns or features in the input image that are associated with various diseases or pests. After training, the network can be utilized to classify new images as diseased or healthy. The classification of raw images is a straightforward process that utilizes the entire image as input to the CNN. However, this approach may be limited by the presence of irrelevant information or noise in the image, which can negatively impact the performance of the network. In order to address this problem, investigators have proposed utilizing a region of interest (ROI) based approach, in which is the model is taught to categorize specific regions of the image that contain the lesion, rather than the entire image. Multi-category classification is another area of research in this field, which involves training CNNs to recognize multiple types of diseases or pests in the same image. This approach can be more challenging than binary classification, as it requires CNNs to learn more complex and diverse patterns in the input images.
The first broad application of CNNs for plant pest and disease detection was the identification of lesions using categorization networks. Current study issues include the categorization of raw pictures, classification following recognition of regions of interest (ROI), and classification of several categories. Utilizing neural structural models, such as CNN, for direct classification in plant pest identification can be a highly effective strategy. CNN is a DL model that is ideally suited for image classification problems since it can automatically learn picture attributes.
To train the network when the team constructed it independently, a tagged collection of photos of ill and healthy plants was required. There must be a variety of pests and illnesses, plant growth phases, and environmental circumstances within the databases. The team can then construct the network architecture and choose relevant parameters based on the specific features of the intended recipient plant pest and disease. Alternately, during transfer learning, you can employ a CNN model that has already been trained and modify it using data from specific plant pest detection tasks. This method is less computationally intensive and requires less labeled data due to the fact that the pre-trained network has already acquired generic characteristics from huge datasets. Notably, transfer learning enables teams to harness the performance of a model trained in some data that were developed using extensive, varied datasets demonstrated to perform well on similar tasks.
Establishing the weight parameters for multi-objective disease and pest classification networks, obtained through binary learning between healthy and infected samples as well as pests, are uniform. A CNN model is designed that integrates basic metadata and allows training on a single multi-crop model to identify 17 diseases across five cultures by utilizing a unified newly suggested model which has ability to handle multiple crops multi-crop model ( Picon et al., 2019 ). The following goals can be accomplished through the use of the proposed model:
1. Achieve more prosperous and stable shared visual characteristics than a single culture.
2. Is unaffected by diseases that cause similar symptoms across cultures.
3. Seamlessly integrates the context for classifying conditional crop diseases.
Experiments show that the proposed model eliminates 71 percent of classification errors and reduces data imbalance, with a balanced data the proposed model boasts an average accuracy rate of 98%, surpassing the performance of other models.
3.5 Identifying lesion locations through neural network analysis
Images are typically processed and labeled using a classification network. However, it is also possible to use a combination of various strategies and methods to determine the location of affected areas and perform pixel-level classification. Some commonly used methods for this purpose include the sliding window approach, the thermal map technique, and the multitasking learning network. These methods involve analyzing the input image and identifying specific regions or areas that correspond to lesions through a systematic and formal analysis process.
The sliding window method is a widely utilized technique for identifying and arranging elements within an image. This method involves moving a small window across the image and analyzing each window using a classification network. This technique is particularly useful for detecting localized features, such as lesions in plant photos, making it a valuable tool. In a study, a CNN classification network incorporating the sliding window method was utilized to develop a system for the identification of plant diseases and pests ( Tianjiao et al., 2019 ). This system incorporates ML, feature fusion, identification, and location regression estimation through the use of sliding window technology. The software demonstrated an ability to identify 70-82% of 29 typical symptoms when used in the field.
The graphic illustrates a temperature chart that illustrates the importance of various regions within an image. The darker the hue, the greater the importance of that region. Specifically, darker tones on the heat map indicate a higher likelihood of lesion detection in plants affected by diseases and pests. In a study conducted by ( Dechant et al., 2017 ), a convolutional neural network (CNN) was trained to generate thermal maps of corn disease images, which were then used to classify the entire image as infected or non-infected. The process of creating a thermal map for a single image takes approximately 2 minutes and requires 2 GB of memory. Identifying a group of three thermal cards for execution, on the other hand, takes less than a second and requires 600 bytes of memory. The results of the study showed that the test data set had an accuracy rate of 98.7%. In a separate study, ( Wiesner-hanks et al., 2019 ) used the thermal map system to accurately identify contour zones for maize diseases with a 96.22% accuracy rate in 2019. This method of detection is highly precise and can identify lesions as small as a few millimeters, making it the most advanced method of aerial plant disease detection to date.
A multitasking learning network is a network that is capable of both categorizing and segmenting plant afflictions and pests. Unlike a pure predictive model, which is only able to categorize images at the image level, multitasking networks add a branch that can accurately locate the affected region of plant diseases. This is achieved by sharing the results of characteristic extraction between the two branches. As a result, the multitasking learning network uses a detection hierarchy to generate precise lesion detection results, which reduces the sampling requirements for the classification network. In a study by ( Shougang et al., 2020 ), a VGCNN model followed by deconvolution (DGVGCNN) was developed to detect afflictions of plant leaves resulting from shadows, obstructions, and luminosity levels. The implementation of deconvolution redirects the CNN classifier’s attention to the precise locations of the afflictions, resulting in a highly robust model with a disease class identification accuracy of 97.81%, a lesion segmentation pixel accuracy of 96.44%, and a disease class recognition accuracy of 98.15%.
Figure 3 presents architecture of the CANet neural network. utilized for plant lesion detection and segmentation. The figure provides a visual representation of the various components and structure of the network, such as the input layer, intermediate hidden layers, and the final output layer. This information is valuable for researchers and practitioners who are interested in understanding the underlying mechanics of the CANet network and how it performs lesion detection and segmentation.
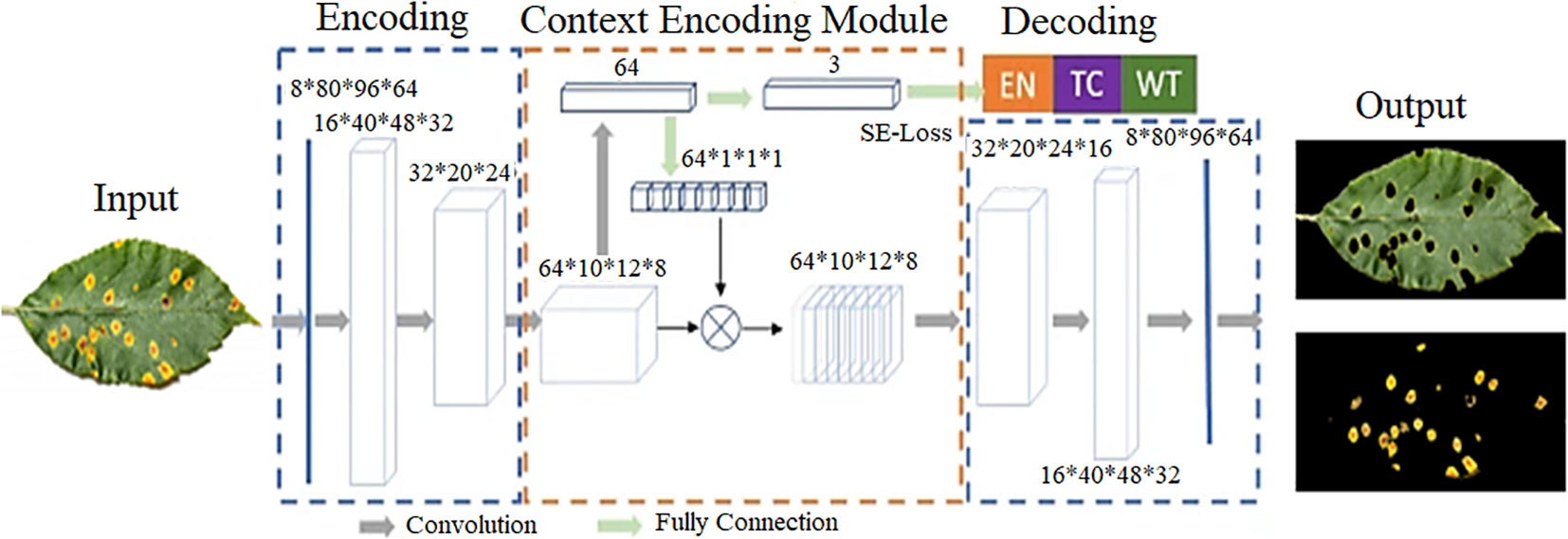
Figure 3 CANet neural network-based disease detection and ROI segmentation ( Shoaib et al., 2022b ).
Table 3 provides a comparison of the pros and cons of various object detection and classification methods for identifying diseases in the leaves of plants. The table compares five methods including Convolutional Neural Networks (CNNs), Transfer learning with CNNs, Multitasking learning networks, Deconvolution-guided VGNet (DGVGNet), and traditional methods such as manual inspection and microscopy. This information is valuable for researchers and practitioners in the area of identifying plant lesions, as it provides a comprehensive comparison of the strengths and limitations of each method, enabling them to make informed decisions about which method is most suitable for their needs. The data presented in Table 3 can act as a guide for future studies and development in the field of plant disease detection.
The research community as a whole has come to acknowledge the utility of taxonomic network systems for the detection of plant pests, and a significant amount of study and investigation is currently being carried out in this field. Table 3 offers a full comparison of the several sub-methods that make up the categorized network system, showing the benefits and drawbacks of each option ( Mohanty et al., 2016 ; Brahimi et al., 2018 ; Garcia and Barbedo, 2019 ). It is essential to keep in mind that the method that will prove to be the most effective will change depending on the particular use case as well as the resources. It should also be mentioned that while this table does illustrate the performance of each approach, it should not be considered to be an exhaustive comparison because the results may differ depending on the particular data sets and environmental conditions that are used.
Table 3 Comparison of pros and cons of various object detection and classification methods for plant leaf disease detection.
3.5.1 Object detection networks for plant lesion detection
Object localization is a fundamental task in computer vision and is closely associated with the traditional detection of plant pests. The objective of this task is to acquire knowledge about the location of objects and their corresponding categories. In recent years, various algorithms for object detection based on DL have been developed. These include single-stage networks such as SSD (W. Liu et al., 2016 ) and YOLO ( Dumitrescu et al., 2022 ; Peng and Wang, 2022 ; Shoaib and Sayed, 2022 ), as well as a networks with multi-stages, like YOLOv1 ( Nasirahmadi et al., 2021 ). These techniques are commonly employed in the identification of plant lesions and pests. The single-stage network makes use of network features to directly forecast the site and classification of blemishes, whereas the two-stage network first generates a candidate box (proposal) with lesions before proceeding to the object detection process.
3.5.2 Pest and plant lesion localization using multi-stage network
Faster R-CNN is a two-part object detection system that uses a common feature extractor to obtain a map of features from an input image. The network then utilizes a Region Proposal Network (RPN) to calculate anchor box confidences and generate proposals. The features maps of the proposed regions are then connected to the ROI pooling layer to enhance the initial detection results and finally determine the location and type of the lesion. This method improves upon traditional structures by incorporating modifications to the feature extractor, anchor ratios, ROI pooling, and loss functions that are tailored to the specific characteristics of plant disease and pest infestation detection. In a study conducted by ( Fuentes et al., 2017a ), the Faster R-CNN was used for the first time to accurately locate tomato diseases and pests infestation in a dataset containing 4800 images of 11 different categories. When using deep feature extractors like VGG-Net and ResNet, the mean average precision (mAP) value was calculated 88.66%.
The YOLOv5 architecture is visually represented in Figure 4 , which depicts its structure and organization. The network comprises three primary components: the input layer, the hidden layers, and the output layer. The input layer is where data is initially fed into the network for processing. The hidden layers are responsible for executing complex computations and transformations on the input data, and their performance plays a critical role in determining the network’s accuracy. The output layer generates final predictions by outputting the bounding boxes and class probabilities for objects detected in the input image. The figure provides detailed labels and annotations to explain how the network’s components interact. This visual representation helps researchers and developers gain a better understanding of the network’s mechanics and identify areas for performance enhancement. Overall, Figure 4 is an essential tool for anyone seeking to deepen their understanding of the YOLOv5 architecture. In 2019, ( Liu and Wang, 2021b ) a modification was suggested for the Faster R-CNN framework to automatically detect beet spot lesion by altering the parameters of the CNN model. A total of 142 images were used for testing and validation, resulting in an overall correct ranking rate of 96.84%. ( Zhou et al., 2019 ) a rapid detection system for rice diseases was proposed by integrating the FCM-Kmeans and YOLOv2 algorithms. The system showed a detection accuracy of 97.33% with a processing time of 0.18s for rice blast, 93.24% accuracy and 0.22s processing time for bacterial blight, and 97.75% accuracy and 0.32s processing time for sheath burn, based on the evaluation of 3010 images. ( Xie et al., 2020 ) proposed the DR-IACNN model based on the faster mechanism to ensure efficiency, a custom dataset is developed that contains the vine leaf lesions (GLDD), and the Faster R-CNN detector employe of a Inception-v2 architecture, the Inception-ResNetv2 architecture. The proposed model showed a mean average precision (mAP) accuracy of 83.7% and a detection rate of 12.09 frames per second. The two-stage detection network was designed to improve the real-time performance and practicality of the detection system. However, it still lacks in terms of speed compared to the speed of one-stage detection model.
Figure 4 YOLOv5 architecture ( Li et al., 2022 ).
3.5.3 One-stage network based plant lesion detection
In recent years, object detection has become an essential tool for diagnosing plant afflictions and pests. YOLO (You Only Look Once) is one of the most widely used object detection techniques. It is a real-time, single-pass object detector that utilizes a single CNN to predict the category and position of objects in an image. Variations of the YOLO algorithm, such as YOLOv2 and YOLOv3, and other various methods have been developed to enhance the accuracy of object recognition while maintaining real-time performance. Another popular object detection technique is SSD (Single Shot MultiBox Detector), which similarly to YOLO, uses a single CNN to predict the type and position of objects in an image. However, SSD makes predictions about the size of objects based on multiple feature maps that are scaled differently, making it better suited for identifying small objects with greater precision than YOLO.
Faster R-CNN is a two-stage object detection system that generates a set of potential object regions using a Region Proposal Network (RPN), and then uses a separate CNN to classify and locate objects within these proposals. Despite being slower than YOLO and SSD, Faster R-CNN has been shown to achieve a higher level of accuracy. When it comes to detecting plant diseases and pests, YOLO, SSD, and Faster R-CNN are all commonly used methods. The choice of algorithm will depend on the specific requirements of the application, such as accuracy, speed, and memory consumption. For real-time applications that prioritize speed, YOLO may be the best option, but for applications that require a higher level of accuracy, SSD and Faster R-CNN may be more suitable.
In this study ( Singh et al., 2020 ), the authors explore the potential of utilizing computer vision techniques for the early and widespread detection of plant diseases. To aid in this effort, a custom dataset, named PlantDoc, was developed for visual plant disease identification. The dataset includes 3,451 data points across 12 plant species and 14 disease categories and was created through a combination of web scraping and human annotation, requiring 352 hours of effort. To demonstrate the effectiveness of the dataset, three plant disease classification models were trained and results showed an improvement in accuracy of up to 29%. The authors believe that this dataset can serve as a valuable resource in the implementation of computer vision methods for plant disease detection.
( Zhang et al., 2019 ) proposed a novel approach to the detection of small agricultural pests by combining an improved version of the YOLOv3 algorithm with a spatial pyramid pooling technique. This method addresses the issue of low recognition accuracy caused by the variable posture and scale of crop pests by applying deconvolution, combining oversampling and convolution operations. This approach allows for the detection of small samples of pests in an image, thus enhancing the accuracy of the detection. The method was evaluated using 20 different groups of pests collected in real-world conditions, resulting in an average identification accuracy of 88.07%. In recent years, many studies have employed detection networks to classify pathogens and pests ( Fuentes et al., 2017a ). It is expected that in the future, more advanced detection models will be utilized for the identification of plant maladies and infestations, as object segmentation networks in computer vision continue to evolve.
In recent times, the detection of plant maladies and infestations has increasingly relied upon the use of two-stage models, which prioritize accuracy. However, there is a growing trend towards the use of single-stage models, which prioritize speed. There has been debate over whether detection networks can replace classification networks in this field. The primary goal of a segmentation network is first to identify the presence of plant maladies and infestations, whereas the goal of a predictive model based on a classification scheme is to categorize these diseases and pests. It is important to note that the visual recognition network provides information on the specific category of diseases and pests that need to be identified. To accurately locate areas of plant disease and pest infestation, detailed annotation is necessary. From this perspective, it may seem that the detection network includes the steps of the classification network. However, it is important to remember that the predetermined categories of plant diseases and pests do not always align with actual results. While the detection network may provide accurate results in different patterns, these patterns may not accurately represent the individuality of specific plant maladies and infestations, and may only indicate the presence of certain kinds of illness and bugs in a specific area. In such cases, the use of a classification network may be necessary. In conclusion, both classification networks and detection networks are important for efficient plant disease and pest detection, but classification networks have more capabilities than detection networks.
3.6 Deep learning-based segmentation network
The segmentation network transforms the task of detecting plant and pest diseases into semantic segmentation, which includes separating lesions from healthy areas. By dividing the lesion’s area in half, it calculates the position, rank, and associated geometric properties (including length, width, surface, contour, center, etc.). Fully convolutional networks include the R-CNN mask ( Lin et al., 2020 ) and completely convolutional networks (FCNs) ( Shelhamer et al., 2017 ).
3.6.1 Fully connected neural network
A complete convolution neural network is used to segment the image’s semantics (FCN). FCN uses convolution to extract and encode the input image features, then deconvolution or oversampling to gradually restore the characteristic image to its original size. FCN is used in almost all semantic segmentation models today. Traditional plant and pest disease segmentation methods are categorized as conventional FCN, U-net ( Navab et al., 2015 ), and SegNet ( Badrinarayanan et al., 2017 ) according to variations in the architecture of the FCN network.
A proposed technique for the segmentation of maize leaf disease employs a fully convolutional neural network (FCN)( Wang and Zhang, 2018 ). The process begins with preprocessing and enhancing the captured image data, followed by the creation of training and test sets for DL. The centralized image is then input into the FCN, where feature maps are generated through multiple layers of convolution, pooling, and activation. The feature map is then up sampled to match the dimensions of the input image. The final step is the restoration of the segmented image’s resolution through the process of deconvolution, resulting in the output of the segmentation process. This method was applied to segment common maize leaf disease images and it was found that the segmentation effect was satisfactory with an accuracy rate exceeding 98%.
The proposed approach employs an improved fully convolutional network (FCN) to precisely segment point regions from crop leaf images with complicated backgrounds ( Wang et al., 2019 ). The strategy addresses the difficulty of reliably identifying sick spots in complicated field situations. The training method of the proposed system employs a collection of crop leaf pictures with healthy and sick sections. The algorithm’s performance is tested using measures such as accuracy and intersectional union ratio (IoU) to determine its ability to effectively partition lesion regions from pictures. The experimental findings demonstrate that the algorithm segments the spot area in complicated backdrop crop leaf images with great precision.
U-Net is a popular CNN architecture for image segmentation tasks. The architecture is named U-Net because it is U-shaped, with encoder and decoder sections connected by a bottleneck ( Shoaib et al., 2022a ). The encoder section of the network consists of a series of convolutional and clustering layers that extract entities from the input image. These features then pass through the bottleneck, where they are up sampled and connected to the feature map from the encoder. This allows the network to use both superficial and fundamental image attributes when making predictions. The decoder part of the network then uses these connected feature maps to generate the final segmentation map. The U-Net architecture is particularly useful for image segmentation tasks because it is able to handle class imbalance problems, where some areas of the image contain more target objects than others.
This paper proposes a semantic segmentation model that uses CNNs to recognize and segment powdery mildew in individual pixel-level images of cucumber lea ( Lin et al., 2019 ). The suggested model obtains an average pixel accuracy of 97.12%, a joint intersection ratio score of 79.54%, and a dice accuracy of 81.54% based on 20 test samples. These results demonstrate that the proposed model outperforms established segmentation techniques such as the gaussian mixture model, random forests, and fuzzy c means. Overall, the proposed model can accurately detect powdery mildew on cucumber leaves at the pixel level, making it a valuable tool for cucumber breeders to assess the severity of powdery mildew.
A novel approach to detect vineyard mildew is proposed, which utilizes DL segmentation on Unmanned Aerial Vehicle (UAV) images ( Kerkech et al., 2022 ). The method involves combining visible and infrared images from two different sensors and using a newly developed image registration technique to align and fuse the information from the two sensors. A fully convolutional neural network is then applied to classify each pixel into different categories, such as shadow, ground, healthy, or symptom. The proposed method achieved an impressive detection rate of 89% at the vine level and 84% at the leaf level, indicating its potential for computer-aided disease detection in vineyards.
3.6.2 Mask regional-CNN
Mask R-CNN is an effective DL model that is perfect for plant pest detection. It is an extension of the Faster R-CNN model and can recognize objects and segment instances ( Permanasari et al., 2022 ). The primary advantage of Mask R-CNN over other models such as YOLO and SSD is its capacity to produce object masks that allow more precise image object location. This is especially beneficial for detecting plant pests, as it enables for more precise identification of afflicted areas. In addition, Mask R-CNN is able to handle overlapping object instances, which is a common issue in plant pest detection due to the presence of several instances of the same pest and disease in a single image. This makes the Mask R-CNN a highly adaptable model that is appropriate for a variety of plant pest identification applications.
In this study ( Stewart et al., 2019 ), an R-CNN based on a masking scheme was utilized to segregate foci of northern plant leaf spots in UAV-captured pictures. The model is trained with a specific data set that recognizes and segments individual lesions in the test set with precision. The average intersectional union ratio (IOU) between the ground reality and the projected lesions was 79.31%, and the average accuracy was 97.24% at a threshold of 60% IOU. In addition, the average accuracy when the IOU threshold ranged from 55% to 90% was 65%. This study illustrates the potential of combining drone technology with advanced instance segmentation techniques based on DL to offer precise, high-throughput quantitative measures of plant diseases.
Using deep CNNs and object detection models, the authors of this paper offer two strategies for tomato disease detection ( Wang et al., 2019 ). These techniques employ two distinct techniques, YOLO and SSD. The YOLO detector is used to categorize tomato disease kinds, while the SSD model is used to classify and separate the ROI-contaminated areas on tomato leaves. Four distinct deep CNNs are merged with two object detection models in order to obtain the optimal model for tomato disease detection. A dataset is generated from the Internet and then split for experimental purposes into training sets, validation sets, and test sets. The experimental findings demonstrate that the proposed approach can accurately and effectively identify eleven tomato diseases and segment contaminated leaf areas.
4 Comparing datasets and evaluating performance
This section starts by providing an overview of the evaluation metrics for DL models, specifically focusing on those that pertain to plant disease and pest detection. It then delves into the various datasets that are relevant to this field, and subsequently, conducts a thorough analysis of the recent DL models that have been proposed for the detection of plant diseases and pests.
4.1 Evaluating plant disease detection using benchmark datasets
The PlantVillage dataset is a compilation of crop photos with labels indicating the presence of various illnesses ( Hughes and Salathé, 2015 ). It features 38,000 photos of 14 distinct crops, including, among others, tomatoes, potatoes, and peppers. The photographs were gathered from many sources, including public databases, research institutions, and individual contributors. The dataset is divided into a training set, a validation set, and a test set, with the training set including the majority of the photos. The scientific community uses this dataset extensively to develop and evaluate DL models for plant disease detection. Figure 5 showcases a selection of images obtained from the PlantVillage dataset, which is a comprehensive dataset containing thousands of images of various plant species. These images depict a wide range of plant conditions, such as healthy plants, plants affected by pests, and plants afflicted by various diseases, which enables researchers and practitioners to gain a comprehensive understanding of the variability in plant growth and development. Moreover, the diverse range of plant species represented in this figure provides an in-depth and realistic representation of the variability in plant types. The images included in this figure capture the nuanced differences in plant morphology, such as leaf shape, color, and texture, which can be useful for developing and validating deep learning models for plant disease detection. The AgriVision collection ( Chiu et al., 2020 ), which contains photos of numerous crops and their diseases, and the Plant Disease Identification dataset, which contains photographs of damaged and healthy plant leaves, are two other significant datasets.

Figure 5 Some random images from plantvillage dataset ( Hughes and Salathé, 2015 ).
Figure 6 showcases a selection of random images obtained from the Agri-Vision dataset. These images depict various crops and their growth conditions, including both healthy and diseased plants. This figure serves as a visual representation of the types of data available in the Agri-Vision dataset, providing insight into the range and diversity of data contained within the dataset. The Crop Disease dataset comprises photos of 14 crops affected by 27 diseases, whereas the Plant-Pathology-2020 dataset provides images of plant leaves damaged by 38 diseases. All of these datasets are widely utilized by the research community and contribute to the creation and evaluation of DL models for plant disease detection.
Figure 6 Some random images from agri-vision dataset ( Chiu et al., 2020 ).
Table 4 provides a summary of benchmark datasets commonly used for plant disease and pest detection. The table includes information on the name of the dataset, a brief description, the type of data contained within the dataset, and the types of diseases and pests covered. This information is valuable for researchers and practitioners who are looking to evaluate or compare their algorithms or models against existing datasets.
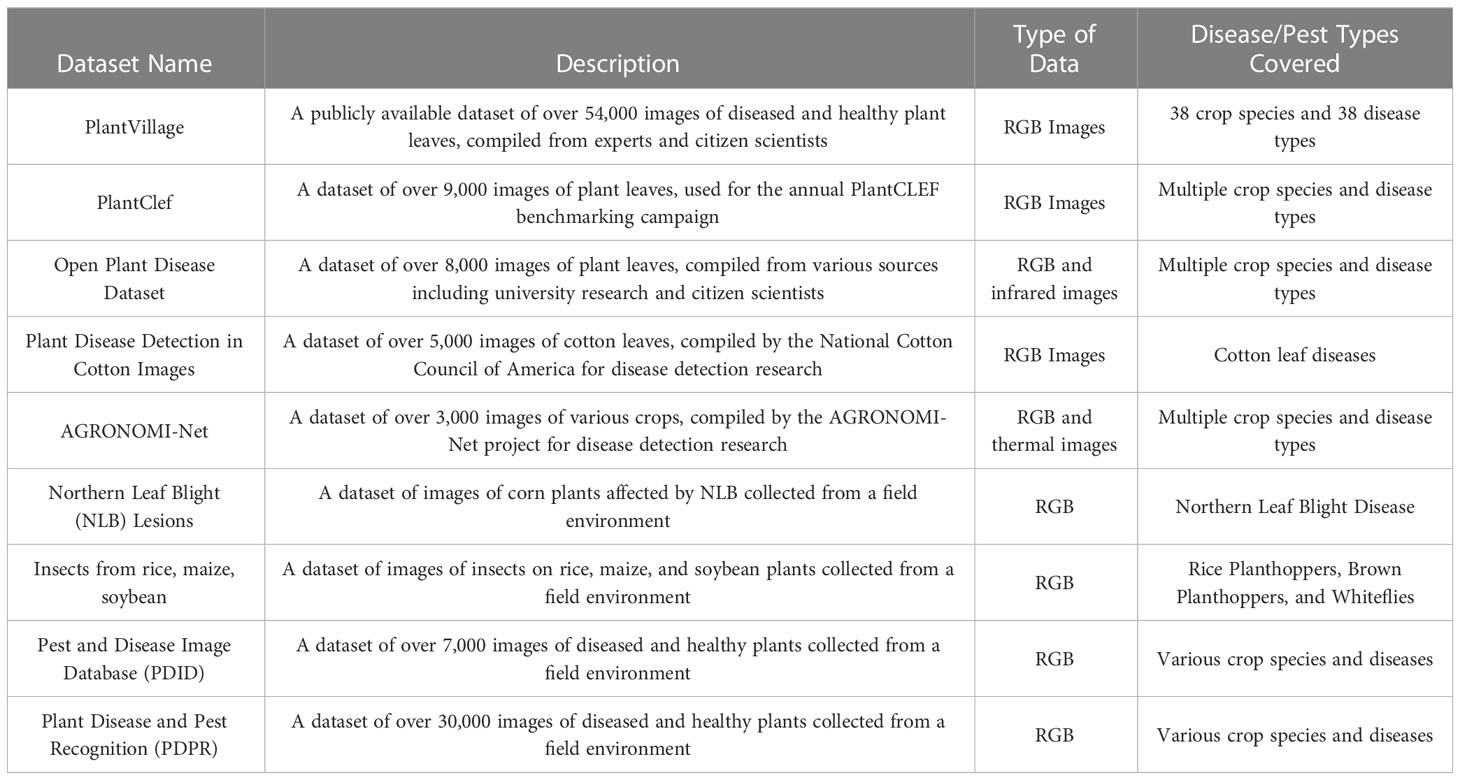
Table 4 Plant disease and pest detection from benchmark datasets.
4.2 Evaluation indices
There are several performance metrics commonly used for evaluating the performance of plant disease classification, detection, and segmentation models. Figure 7 displays an example of a confusion matrix, a widely used evaluation metric in machine learning. The matrix represents the results of a classification algorithm, where each row represents the predicted class of a given sample and each column represents the actual class of that sample. The entries in the matrix show the number of samples that have been correctly or incorrectly classified. By examining the entries in the confusion matrix, it is possible to gain insight into the performance of the classification algorithm and identify areas for improvement.
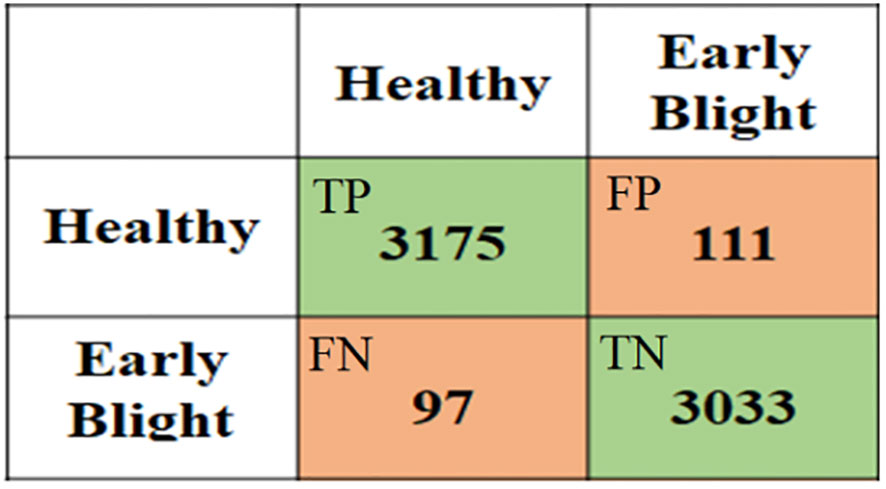
Figure 7 An example of a confusion matrix where the rows show the predicted results while columns represent actual classes.
Accuracy: This is the proportion of correctly classified instances out of the total number of instances. Mathematically, it is represented as:
Precision: This is the proportion of correctly classified positive instances out of the total number of predicted positive instances. Mathematically, it is represented as:
Recall (Sensitivity): This is the proportion of correctly classified positive instances out of the total number of actual positive instances. Mathematically, it is represented as:
F1 Score: This is the harmonic mean of precision and recall. Mathematically, it is represented as:
Intersection over Union (IoU): This is used to evaluate the performance of segmentation models. It is the ratio of the area of intersection of the predicted segmentation and the ground truth segmentation to the area of the union of the two. Mathematically, it is represented as:
Dice coefficient: This is another metric used for evaluating segmentation performance. It is a measure of the similarity between the predicted segmentation and the ground truth segmentation, and it ranges from 0 to 1. Mathematically, it is represented as:
Jaccard index: This is another metric used for evaluating segmentation performance. It is the ratio of the area of intersection of the predicted segmentation and the ground truth segmentation to the area of the union of the two. Mathematically, it is represented as:
Receiver Operating Characteristic: This curve is a graphical representation of the performance of a binary classifier system. Figure 8 presents an example of a performance comparison between three models using a receiver operating characteristic (ROC) curve. The ROC curve is a widely used evaluation metric in machine learning that graphically summarizes the performance of a binary classifier by plotting the true positive rate against the false positive rate for different classification thresholds. The ROC curve provides a visual representation of the trade-off between the false positive rate and true positive rate, allowing practitioners to compare the performance of different models at different operating points. It plots the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings. The TPR, also known as the sensitivity, recall or hit rate, is the number of true positive predictions divided by the number of actual positive cases. The FPR, also known as the fall-out or probability of false alarm, is the number of false positive predictions divided by the number of actual negative cases. The ROC curve can be mathematically represented as TPR = (TP)/(TP + FN) and FPR = (FP)/(FP + TN), where TP, FP, TN, and FN are true positives, false positives, true negatives, and false negatives, respectively. The area under the ROC curve (AUC) is a measure of the classifier’s performance, with a value of 1 indicating perfect performance and a value of 0.5 indicating no better than random.
Figure 8 An example of performance comparison between three models using the ROC curve ( Shoaib et al., 2022a ).
Area Under the Curve: This AUC is also a performance measure used to evaluate the performance of the binary classifier. It is derived by integrating the true positive rate (TPR) relative to the false positive rate (FPR) overall thresholds. TPR is determined by dividing the number of true positives by the total number of true positive instances (TP + FN), whereas FPR is determined by dividing the number of false positives by the total number of true negative cases (FP + TN). AUC goes from 0 to 1, where 1 corresponds to a perfect classifier and 0.5 corresponds to a random classifier. A greater AUC value suggests superior classification ability.
4.3 Performance comparison of existing algorithms
This article examines in depth the most recent developments in DL-based plant pest identification. The papers examined in this article, published between 2015 and 2022, focus on the detection, classification, and segmentation of plant pests and lesions using ML and DL approaches. This research employs several methodologies, including image processing, feature extraction, and classifier creation. In addition, DL models, namely CNNs, have been widely applied to accurately detect and categorize plant illnesses. This article addresses the problems and limits of utilizing ML and DL algorithms for plant lesions identification, including data availability, image quality, and subtle differences between healthy and diseased plants. This paper also examines the current state of practical applications of ML and DL techniques in plant abnormal region detection and provides viable solutions to address the obstacles and limits of these technologies.
The research covered in this article indicates that the employment of ML and DL approaches enhances the accuracy and efficiency of plant lesion detection greatly. The most prevalent evaluation criteria are mean accuracy (mAP), F1 score, and frames per second (FPS). However, a gap still exists between the intricacy of the images of infectious maladies and infestations utilized in this study and the usage of mobile devices to identify pest and lesions infestations in the field in real-time. This paper is a valuable resource for plant lesions detection researchers, practitioners, and industry experts. It provides a comprehensive understanding of the current state of research utilizing ML and DL techniques for plant lesions detection, highlights the benefits and limitations of these methods, and proposes potential solutions to overcome the challenges of their implementation. In addition, the need for larger and more intricate experimental data sets was identified as a subject for further investigation.
5 Challenges in existing systems
5.1 overcoming small dataset challenge.
Using data augmentation techniques to fictitiously expand the dataset is one method. Another strategy is to use knowledge from models that have already been trained on bigger data sets to smaller data sets. The third approach successfully addresses the small sample problem by combining the first two approaches. Despite these achievements, a significant obstacle in the field of DL-based plant pest identification is still the limited dataset problem. Future research should therefore concentrate on creating new tools and techniques to successfully address this issue and enhance the functionality of DL models in this domain.
5.2 Plant image amplification for lesions segmentation
In recent years, data amplification technology has been utilized extensively in the field of plant pest detection in order to circumvent the issue of small data set size. These techniques involve the use of image manipulation operations including mirroring, translation, shearing, scaling, and contrast alteration in order to create additional training examples for a DL model. In order to enrich tiny datasets, generative adversarial networks (GANs) ( Goodfellow et al., 2020 ) and automated encoders ( Pu et al., 2016 ) were also utilized to generate fresh, diverse samples. It has been demonstrated that these strategies considerably enhance the performance of DL models for plant pest detection. It is essential to emphasize, however, that the efficacy of these strategies is contingent on the quality and diversity of the original dataset. Additionally, the produced samples must be thoroughly analyzed to confirm their suitability for DL model training. Data amplification, synthesis, and generative approaches are crucial components of plant pest detection model training using DL.
5.3 Transfer learning for plant disease and pest detection
Transfer learning is a technique that applies models that have been trained on large, generic datasets to more specific tasks with fewer data. This method is especially beneficial in the field of plant pest detection, where annotated data is frequently sparse. Pretrained models can be customized for specific localized plant pest and abnormality detection tasks by refining parameters or fine-tuning certain components. Transfer learning can increase model performance and minimize model development expenses, according to studies. For example, ( Oppenheim et al., 2019 ) used the VGG network to recognize natural light images of contaminated potatoes of various sizes, colors, and forms. ( Too et al., 2019 ) discovered that as the number of iterations grew, the accuracy of dense nets improved when employing fine and contrast parameters. In addition, ( Chen et al., 2020 ) demonstrate that transfer learning can accurately diagnose rice lesions photos in complicated situations with an average accuracy of 94 percent, exceeding standard training.
5.4 Optimizing network structure for plant lesion segmentation
A properly designed array structure can greatly minimize the number of samples required for plant pest and lesions segmentation. Utilizing several color channels, merging depth-separate convolution, and adding starting structures are some of the strategies employed by researchers to increase feature extraction. Specifically, Identification of plant leaf diseases using RGB pictures and a convolutional neural network with three channels (TCCNN) in ( Zhang et al., 2019 ). An enhanced CNN approach that uses deep separable convolution to detect illnesses in grapevine leaves is proposed in ( Liu et al., 2020 ), with 94.35% accuracy and faster convergence than classic ResNet and GoogLeNet structures. These examples illustrate the significance of examining network patterns for detecting plant pests and diseases with limited sample numbers.
5.5 Small-size lesions in early identification
The primary role of the attention mechanism is to pinpoint the area of interest and swiftly discard unnecessary data. A weighted sum approach with weighted coefficients can be used to separate the features and reduce background noise in plant and pest images by analyzing the images’ features. Specifically, the Attention Mechanism module can build a new noise reduction fusion function using the Softmax function by capturing the prominent image, isolating the item from the context, and utilizing and fusing the feature image with the original feature image. The attention mechanism can efficiently choose data and assign enhanced resources to the ROIs, allowing for additional precise identification of minor lesions during the early stages of pest infestations and diseases. Numerous research, such as ( Karthik et al., 2020 ) have demonstrated the efficacy of the attention based prediction system. On the industrial village dataset, the network residual attention mechanism was evaluated with an overall accuracy of 98%. In addition, to improve the precision of tiny lesion detection, research can concentrate on creating more robust preprocessing algorithms to reduce background noise and enhance picture resolution. This may involve techniques such as picture enhancement, image denoising, and image super-resolution.
5.6 Fine-grained identification
The identification of plant diseases and pests is a challenging task that is often made more complex by variations in the visual characteristics of affected plants. These variations can be attributed to external factors such as uneven lighting, extensive occlusion, and fuzzy details ( Wang et al., 2017 ). Furthermore, variations in the presence of illness and the growth of a pest can lead to subtle differences in the characterization of the same diseases and pests in different regions, resulting in “intra-class distinctions” ( Barbedo, 2018 ). Additionally, there is a problem of “inter-class resemblance,” which arises from similarities in the biological morphology and lifestyles of subclasses of diseases and pests, making it difficult for plant pathologists to differentiate between them.
In actual agricultural settings, the presence of background disturbances might make it harder to detect plant pests and diseases ( Garcia and Barbedo, 2018 ). Environment complexity and interactions with other items can further complicate the detecting procedure. It is essential to highlight, however, that images obtained under controlled conditions may not truly depict the difficulties of spotting pests and illnesses in their natural habitats. Despite advancements in DL techniques, identifying pests and diseases in real-world contexts remains a technological issue with accuracy and robustness constraints. Current research focuses mostly on the fine-grained identification of individual pest populations, and it is challenging to apply these methods to mobile, intelligent agricultural equipment for large-scale identification. Therefore, additional study is required to address these obstacles and enhance the effectiveness of agricultural decision management.
5.7 Low and high illumination problem
In the past, researchers captured photos of plant pests and illnesses using indoor lightboxes ( Martinelli et al., 2015 ). Despite the fact that this method efficiently eliminates the impacts of outdoor lighting, hence simplifying picture processing, it is essential to remember that photographs captured under natural lighting circumstances might vary significantly. The dynamic nature of natural light and the limited range of the camera’s dynamic light source might create color distortion if the camera settings are not appropriately adjusted. Moreover, the visual attributes of plant illnesses and infestations may be impacted by factors such as viewing angle and distance, offering a formidable challenge to visual recognition algorithms. This emphasizes the significance of addressing light conditions and image capture techniques when researching pests and plant diseases, as these factors can significantly impact the accuracy and dependability of results.
5.8 Challenges posed by obstruction
Currently, the majority of scientists tend to concentrate on detecting plant pests and diseases in particular ecosystems, rather than addressing the setting as a whole. Frequently, they directly intercept areas of interest in the gathered photos without completely resolving the occlusion issue. This results in low recognition accuracy and restricted applicability. There are numerous types of occlusions, including differences in leaf location, branches, external lighting, and hybrid designs. These occlusion issues are ubiquitous in the natural environment, where a lack of distinguishing characteristics and overlapping noise makes it difficult to identify plant pests and illnesses. In addition, varying degrees of occlusion may have varying effects on the recognition process, leading to errors or missed detections. Some researchers have found it challenging to identify plant pests and diseases under extreme conditions, such as in the shadow, despite recent breakthroughs in DL algorithms ( Liu and Wang, 2020 ; Liu and Wang, 2021a ). However, in recent years, a solid foundation has been established for plant utilization and pest identification in actual situations.
To improve the performance of plant pest and disease detection, it is necessary to increase the originality and efficiency of the underlying architecture, which must be improved for optimal results of lightweight network topologies. The difficulty of constructing a core framework is frequently reliant on the performance of the hardware system. Consequently, optimizing the underlying framework is crucial for enhancing efficiency and performance. Moreover, processing blockage might be unanticipated and difficult to anticipate. Therefore, it is essential to lower the complexity of model formation while simultaneously enhancing GAN exploration and preserving detection precision. GANs have the capacity to manage postural shifts and turbulent settings well. However, GAN architecture is still in its infancy and prone to issues during the learning and training phase. To aid in the evaluation of the model’s efficacy, it is essential to do additional research on the network’s outcomes.
5.9 Challenges in detection efficiency
DL algorithms have proven more effective than conventional approaches, although they are computationally intensive. This causes slower inspections and challenges in satisfying real-time requirements, particularly when a high level of detection precision is required. Frequently, in order to resolve this issue, it is required to minimize the amount of data used, which might result in poor planning and erroneous or lost identification. Therefore, it is vital to create an accurate and effective algorithm for threat identification. In agricultural applications, the process of detecting pests and illnesses using DL approaches requires three main steps: data labeling, model training, and model inference. The model inference is particularly applicable to agricultural applications in real-time. However, it should be highlighted that the majority of current mechanisms for disease and bug detection in plants rely on accurate identification, while less emphasis has been paid to the dependability of model inference. For instance, the author of ( Kc et al., 2019 ) employs an ensemble convolutional structural framework to identify plant foliar diseases in order to improve the efficiency of the model calculation process and satisfy real agricultural needs. This approach was compared to various different models, and the decreased MobileNet classification accuracy was 92.12%, with parameters that were 31 times lower than VGG and 6 times lower than MobileNet. This demonstrates that real-time crop disease diagnostics on mobile devices with limited resources strike a solid balance between speed and accuracy.
6 Discussion
6.1 datasets for identifying plant diseases and pests.
The advancement of DL technology has greatly contributed to the improvement of Identifying and managing infestations in crops and plants. Theoretical developments in image identification mechanisms have paved the way for identifying complex diseases and pests. However, it should be noted that the majority of research in this field is limited to laboratory studies and relies heavily on photographs of plant diseases and pests that have been collected. Previous research often focused on identifying specific features such as disease spots, insect appearance, and leaf identification. However, it is important to consider that plant growth is cyclical, consistent, seasonal, and regional in nature. Therefore, it is crucial to gather sample images from various stages of plant growth, different seasons, and regions to ensure a more comprehensive understanding of plant diseases and pests. This will improve the robustness and generalization of the model.
It is essential to keep in mind that the properties of plant diseases or insects which may vary in various phases of crop development. Moreover, photos of different plant species may change by location. Consequently, the majority of current research findings may not be universally relevant. Even if the recognition rate of a single test is high, the reliability of data collected at other times or locations cannot be confirmed. Much of the present study has concentrated on images in the visible spectrum, but it is crucial to remember that electromagnetic waves generate vast amounts of data outside of the visible spectrum. It is necessary to merge data from multiple sources, such as visible, near-infrared, and multispectral, to generate a comprehensive dataset on plant diseases. Future studies will emphasize the use of multi-dimensional concatenation (fusion) techniques to gather and recognize information on plant insects. It should also be highlighted that a database containing photographs of many wild plant pests and illnesses is currently in the process of being compiled. Future studies can use wearable automatic field spore traps, drone aerial photography systems, agricultural Internet of Things monitoring devices, etc. to identify wide regions of farmland, compensating for the absence of randomness in prior studies’ image samples. Improve the overall performance of the algorithm by ensuring the dataset is complete and accurate.
6.2 Pre-emptive detection of plant diseases and pests
Early Identifying the various forms of plant diseases and pests can be a difficult task. due to the fact that symptoms are not always apparent, either through visual inspection or computer analysis. In terms of research and necessity, however, early identification is essential since it helps prevent and control the spread and growth of pests and diseases. Recording photographs under favorable lighting conditions, such as sunny weather, enhance image quality, but capturing images on overcast days complicates preprocessing and decreases identification accuracy. In addition, it might be difficult to understand even high-resolution photos during the first phases of plant pests and diseases. It is necessary to incorporate meteorological and plant health data, such as temperature and humidity, to efficiently identify and predict pests and diseases. Rarely has this technique been utilized to diagnose early plant pests and diseases.
6.3 Neural network learning and development
Manual pest and disease testing are tough since it is difficult to sample for all pests and diseases, and oftentimes only accurate data are available (positive samples). However, the majority of existing systems for plant pest and disease identification utilizing DL are based on supervised learning, which involves the time-consuming collection of huge labeled datasets. Consequently, it is worthwhile to research methods of unsupervised learning. In addition, DL can be a “black box” with little explanatory power, necessitating the labeling of many learning samples for end-to-end learning. In order to assist training and network learning, it may be advantageous to combine past knowledge of brain-like computers with human visual cognitive models.
However, depth models demand a great deal of memory and testing time, making them inappropriate for mobile platforms with limited resources. Therefore, it is necessary to find solutions to reduce model complexity and speed without sacrificing precision. Choosing appropriate hyperparameters, such as learning rate, filter size, step size, and number, has proven to be a significant challenge when applying DL models to new tasks. These hyperparameters have high internal dependencies, so even small changes can have a substantial effect on the final training results.
6.4 Cross-disciplinary study
Theories such as scientific evidence and agronomic plant defenses will be merged to produce more effective field diagnostic models for crop growth and disease identification. Using this technology, plant and pest diseases can be diagnosed with greater speed and precision. In the future, it will be important to shift beyond simple surface image analysis to determine the underlying mechanisms by which pests and diseases occur, together with a full understanding of crop growth patterns, environmental conditions, and other pertinent elements. DL approaches have been demonstrated to address complicated problems that regular image processing and ML methods cannot. Despite the fact that the practical implementation of this technology is still in its infancy, it has enormous development and application potential. To reach this potential, specialists from a variety of fields, such as agriculture and plant protection, must combine their knowledge and experience with DL algorithms and models. In addition, the outcomes of this study will need to be incorporated into agricultural gear and equipment to accomplish the desired theoretical effect.
6.5 Deep learning for plant stress phenotyping: Trends and perspectives
DL and ML technologies are successful in detecting and analyzing lesions from severe abiotic stresses, such as drought. In the past decade, global crop production losses due to drought have totaled approximately $30 billion ( Agarwal et al., 2020 ). In 2012, a severe drought impacted 80% of agricultural land in the US, resulting in over two-thirds of counties being declared disaster areas. According to FAO (UN) reports, drought is the primary cause of agricultural production loss. Drought stress causes 34% of crop and livestock production loss in LDCs and LMICs, costing 37 billion USD. Agriculture sustains 82% of all drought impact. Understanding how plants adapt to stress, especially drought, is essential for securing crop yields in agriculture. DL and ML approaches are therefore a major advance in the field of plant stress biology. ML and DL can be used to categorize plant stress phenotyping problems into four categories: identification, classification, quantification, and prediction ( Singh et al., 2020 ). These categories represent a progression from simple feature extraction to increasingly more complex information extraction from images. Identification involves detecting specific stress types, such as sudden death syndrome in soybeans or rust in wheat. Classification uses ML to categorize the images based on stress symptoms and signatures, dividing the visual data into distinct stress classes, such as low, medium, or high stress categories. The final category, prediction, involves anticipating plant stress before visible symptoms appear, providing a timely and cost-effective way to control stress and advancing precision and prescriptive agriculture.
6.6 Limitations of this study
The study presented in this paper has some limitations that are attributed to its research methodology. Firstly, the study’s scope is confined to publications from 2015 to 2022, implying that recent developments in plant disease detection may not be covered. Moreover, the review does not encompass an all-inclusive list of Machine Learning (ML) and Deep Learning (DL) techniques for plant disease detection. Nevertheless, the study provides an overview of the most commonly used techniques, their advantages, limitations, and probable solutions to overcome implementation challenges. Finally, the study fails to include an extensive examination of the economic and environmental impacts of ML and DL techniques on plant disease detection. Hence, additional research is necessary to scrutinize the potential benefits and disadvantages of these techniques regarding production losses and resource utilization.
6.7 Practical implications of study
The practical implications of our research include:
● Improved plant disease detection: Our research highlights the effectiveness of using ML and DL techniques for plant disease detection, which can help improve the accuracy and efficiency of disease detection compared to traditional manual methods. By adopting these advanced technologies, farmers and plant disease specialists can detect diseases at an early stage, preventing further spread and reducing the risk of crop losses.
● Development of generalizable models: Our research emphasizes the need for developing generalizable models that can work for different plant species and diseases. The development of such models can save time and effort for researchers and practitioners, making it easier to detect and classify plant diseases in various settings.
● Accessible datasets for training and evaluation: The research emphasizes the need for more publicly available datasets for training and evaluating ML and DL models for plant disease detection. The availability of such datasets can help researchers and practitioners develop more accurate and robust models, enhancing the performance of disease detection systems.
● Potential for cost reduction: The use of ML and DL techniques in plant disease detection can reduce the need for manual labor and the cost of plant disease detection. This can be especially useful for farmers and small-scale agricultural operations who may not have access to expensive equipment or specialized expertise.
● Transferable knowledge to other fields: Our research also has the potential to inform research and development in other fields, such as medical imaging and remote sensing. The techniques and methodologies used in plant disease detection can be applied to other fields, providing insights into the potential applications of ML and DL in various domains.
7 Conclusions
The DL and ML technologies have greatly improved the detection and management of crop and plant infestations. Advances in image recognition have made it possible to identify complicated diseases and pests. However, most research in this area is limited to lab-based studies and heavily relies on collected plant disease and pest photos. To enhance the robustness and generalization of the model, it’s important to gather images from various plant growth stages, seasons, and regions. Early identification of plant diseases and pests is crucial in preventing and controlling their spread and growth, thus incorporating meteorological and plant health data, such as temperature and humidity, is necessary for efficient identification and prediction. Unsupervised learning and integrating past knowledge of brain-like computers with human visual cognition can aid in DL model training and network learning. Achieving the full potential of this technology requires collaboration between specialists from agriculture and plant protection, combining their knowledge and experience with DL algorithms and models, and integrating the results into farming equipment. The paper explores the recent progress in using ML and DL techniques for plant disease identification, based on publications from 2015 to 2022. It demonstrates the benefits of these techniques in increasing the accuracy and efficiency of disease detection, but also acknowledges the challenges, such as data availability, imaging quality, and distinguishing healthy from diseased plants. The study finds that the use of DL and ML has significantly improved the ability to identify and detect plant diseases. The novelty of this research lies in its comprehensive analysis of the recent developments in using ML and DL techniques for plant disease identification, along with proposed solutions to address the challenges and limitations associated with their implementation. By exploring the benefits and drawbacks of various methods, and offering valuable insights for researchers and industry professionals, this study contributes to the advancement of plant disease detection and prevention.
Authors contributions
MS, BS, SE-S, AA, AU, FayA, TG, TH, and FarA performed the data analysis, conceptualized this study, designed the experimental plan, conducted experiments, wrote the original draft, revised the manuscript. All authors contributed to the article and approved the submitted version.
AA acknowledges project CAFTA, funded by the Bulgarian National Science Fund. TG acknowledges the European Union’s Horizon 2020 research and innovation programme, project PlantaSYST (SGA-CSA No. 739582 under FPA No. 664620) and the BG05M2OP001-1.003-001-C01 project, financed by the European Regional Development Fund through the Bulgarian’ Operational Programme Science and Education for Smart Growth. This research work was also supported by the Cluster grant R20143 of Zayed University, UAE.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abadi, M. (2016). “TensorFlow: learning functions at scale,” in Proceedings of the 21st ACM SIGPLAN International Conference on Functional Programming, Nara Japan, September 18 - 24, 2016. (Japan: ACM digital library), 1–1. doi: 10.1145/2951913.2976746
CrossRef Full Text | Google Scholar
Agarwal, M., Singh, A., Arjaria, S., Sinha, A., Gupta, S. (2020). ToLeD: Tomato leaf disease detection using convolution neural network. Proc. Comput. Sci. 167 (2019), 293–301. doi: 10.1016/j.procs.2020.03.225
Akbar, M., Ullah, M., Shah, B., Khan, R. U., Hussain, T., Ali, F., et al. (2022). An effective deep learning approach for the classification of bacteriosis in peach leave. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1064854
PubMed Abstract | CrossRef Full Text | Google Scholar
Alzubaidi, L., Zhang, J., Humaidi, A. J., Al-Dujaili, A., Duan, Y., Al-Shamma, O., et al. (2021). Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J. Big Data 8, 1–74. doi: 10.1186/s40537-021-00444-8
Anjna, Sood, M., Singh, P. K. (2020). Hybrid system for detection and classification of plant disease using qualitative texture features analysis. Proc. Comput. Sci. 167 (2019), 1056–1065. doi: 10.1016/j.procs.2020.03.404
Antonellis, G., Gavras, A. G., Panagiotou, M., Kutter, B. L., Guerrini, G., Sander, A. C., et al. (2015). Shake table test of Large-scale bridge columns supported on rocking shallow foundations. J. Geotechnical Geoenvironmental Eng. 12, 04015009. doi: 10.1061/(ASCE)GT.1943-5606.0001284
Arcaini, P., Bombarda, A., Bonfanti, S., Gargantini, A. (2020). “Dealing with robustness of convolutional neural networks for image classification,” in Proceedings - 2020 IEEE International Conference on Artificial Intelligence Testing, AITest 2020. (Oxford, UK: IEEE), 7–14. doi: 10.1109/AITEST49225.2020.00009
Badrinarayanan, V., Kendall, A., Cipolla, R. (2017). Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39 (12), 2481–2495. doi: 10.1109/TPAMI.2016.2644615
Bahrampour, S., Ramakrishnan, N., Schott, L., Shah, M. (2015). Comparative study of caffe, neon, theano, and torch for deep learning. arXiv preprint arXiv:1511.06435 132, 1–9. doi: 10.48550/arXiv.1511.06435
Barbedo, J. G. A. (2018). ScienceDirect factors influencing the use of deep learning for plant disease recognition. Biosyst. Eng. 172, 84–91. doi: 10.1016/j.biosystemseng.2018.05.013
Brahimi, M., Arsenovic, M., Laraba, S., Sladojevic, S., Boukhalfa, K., Moussaoui, A. (2018). Deep learning for plant diseases: detection and saliency map visualisation. Hum. Mach. Learning: Visible Explainable Trustworthy Transparent 6, 93–117. doi: 10.1007/978-3-319-90403-0_6
Cedric, L. S., Adoni, W. Y. H., Aworka, R., Zoueu, J. T., Mutombo, F. K., Krichen, M., et al. (2022). Crops yield prediction based on machine learning models: Case of West African countries. Smart Agric. Technol. 2 (March), 100049. doi: 10.1016/j.atech.2022.100049
Chen, J., Chen, J., Zhang, D., Sun, Y., Nanehkaran, Y. A. (2020). Using deep transfer learning for image-based plant disease identi fi cation. Comput. Electron. Agric. 173 (April), 105393. doi: 10.1016/j.compag.2020.105393
Chiu, M. T., Xu, X., Wei, Y., Huang, Z., Schwing, A., Brunner, R., et al. (2020). Agriculture-Vision : A Large Aerial Image Database for Agricultural Pattern Analysis. in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). (WA, USA: IEEE), 2825–2835. doi: 10.1109/CVPR42600.2020.00290
Chung, Y., Ahn, S., Yang, J., Lee, J. (2017). Comparison of deep learning frameworks: about theano, tensorflow, and cognitive toolkit. J. Intell. Inf. Syst. 23 (2), 1–17. doi: 10.13088/jiis.2020.26.4.027
Dechant, C., Wiesner-hanks, T., Chen, S., Stewart, E. L., Yosinski, J., Gore, M. A., et al. (2017). Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology 107, 1426–1432. doi: 10.1094/PHYTO-11-16-0417-R
Dhaka, V. S., Meena, S. V., Rani, G., Sinwar, D., Ijaz, M. F., Woźniak, M. (2021). A survey of deep convolutional neural networks applied for prediction of plant leaf diseases. Sensors 21 (14), 4749. doi: 10.3390/s21144749
Dillon, J. V., Langmore, I., Tran, D., Brevdo, E., Vasudevan, S., Moore, D., et al. (2017). Tensorflow distributions. arXiv preprint arXiv:1711.10604 , 1–10. doi: 10.48550/arXiv.1711.10604
Domingues, T., Brandão, T., Ferreira, J. C. (2022). Machine learning for detection and prediction of crop diseases and pests: A comprehensive survey. Agriculture 12 (9), 1350. doi: 10.3390/agriculture12091350
Drenkow, N., Sani, N., Shpitser, I., Unberath, M. (2021). Robustness in deep learning for computer vision: mind the gap? arXiv preprint arXiv:2112.00639 , 1–23. doi: 10.48550/arXiv.2112.00639
Dumitrescu, F., Boiangiu, C. A., Voncila, M. L. (2022). Fast and robust people detection in RGB images. Appl. Sci. (Switzerland) 12 (3), 1–24. doi: 10.3390/app12031225
Fuentes, A., Lee, J., Lee, Y., Yoon, S., Park, D. S. (2017a). “Anomaly detection of plant diseases and insects using convolutional neural networks,” in Proceedings of the International Society for Ecological Modelling Global Conference. (Jeju, South Korea: Elsevier).
Google Scholar
Fuentes, A., Yoon, S., Kim, S. C., Park, D. S. (2017b). A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors (Switzerland) 17 (9), 1–21. doi: 10.3390/s17092022
Garcia, J., Barbedo, A. (2018). Impact of dataset size and variety on the e ff ectiveness of deep learning and transfer learning for plant disease classi fi cation. Comput. Electron. Agric. 153 (July), 46–53. doi: 10.1016/j.compag.2018.08.013
Garcia, J., Barbedo, A. (2019). ScienceDirect plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 180 (2016), 96–107. doi: 10.1016/j.biosystemseng.2019.02.002
Genaev, M. A., Skolotneva, E. S., Gultyaeva, E. I., Orlova, E. A., Bechtold, N. P., Afonnikov, D. A.. (2021). Image-based wheat fungi diseases identification by deep learning. Plants 10 (8), 1–21. doi: 10.3390/plants10081500
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2020). Generative adversarial networks. Commun. ACM 63 (11), 139–144. doi: 10.48550/arXiv.1406.2661
Hasan, H., Shafri, H. Z. M., Habshi, M. (2019). “A comparison between support vector machine (SVM) and convolutional neural network (CNN) models for hyperspectral image classification,” in IOP Conference Series: Earth and Environmental Science, (Kula Lumpur, Malysia: IOP science) 357. doi: 10.1088/1755-1315/357/1/012035
Hasan, R. I., Yusuf, S. M., Alzubaidi, L. (2020). Review of the state of the art of deep learning for plant Diseases : A broad analysis and discussion. Plants 9, 1–25. doi: 10.3390/plants9101302
Huang, G., Liu, Z., van der Maaten, L., Weinberger, K. Q. (2017). “Densely connected convolutional networks,” in Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, 2017-January. (USA: IEEE) 2261–2269. doi: 10.1109/CVPR.2017.243
Hughes, D., Salathé, M. (2015). An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv preprint arXiv:1511.08060 , 1–7. doi: 10.48550/arXiv.1511.0806
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., et al. (2014). Caffe : Convolutional architecture for fast feature embedding ∗ categories and subject descriptors. In Proceedings of the 22nd ACM international conference on Multimedia. (Florida, USA) 13, 675–678. doi: 10.48550/arXiv.1408.5093
Karthik, R., Hariharan, M., Anand, S., Mathikshara, P., Johnson, A., Menaka, R. (2020). Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. 86, 105933. doi: 10.1016/j.asoc.2019.105933
Kaur, L., Sharma, S. G. (2021). Identification of plant diseases and distinct approaches for their management. Bull. Natl. Res. Centre 45 (1), 1–10. doi: 10.1186/s42269-021-00627-6
Kc, K., Yin, Z., Wu, M., Wu, Z. (2019). Depthwise separable convolution architectures for plant disease classi fi cation. Comput. Electron. Agric. 165 (December 2018), 104948. doi: 10.1016/j.compag.2019.104948
Kerkech, M., Hafiane, A., Canals, R. (2020). Vine disease detection in UAV multispectral images using optimized image registration and deep learning segmentation approach. Comput. Electron. Agric. 174, 105446. doi: 10.1016/j.compag.2020.105446
Khan, R. U., Khan, K., Albattah, W., Qamar, A. M. (2021). Image-based detection of plant diseases: from classical machine learning to deep learning journey. Wireless Commun. Mobile Computing 2021, 1–13. doi: 10.1155/2021/5541859
Komar, M., Yakobchuk, P., Golovko, V., Dorosh, V., Sachenko, A. (2018). “Deep neural network for image recognition based on the caffe framework,” in 2018 IEEE Second International Conference on Data Stream Mining & Processing (DSMP), (Lviv, Ukraine: IEEE) 102–106. doi: 10.1109/DSMP.2018.8478621
Kurmi, Y., Gangwar, S. (2022). A leaf image localization based algorithm for different crops disease classification. Inf. Process. Agric. 9 (3), 456–474. doi: 10.1016/j.inpa.2021.03.001
Lee, W. H., Ozger, M., Challita, U., Sung, K. W. (2021). Noise learning-based denoising autoencoder. IEEE Commun. Lett. 25 (9), 2983–2987. doi: 10.1109/LCOMM.2021.3091800
Li, J., Liu, C., Lu, X., Wu, B. (2022). CME-YOLOv5: An efficient object detection network for densely spaced fish and small targets. Water (Switzerland) 14 (15), 1–12. doi: 10.3390/w14152412
Lin, K., Gong, L., Huang, Y., Liu, C., Pan, J. (2019). Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00155
Lin, T. Y., Goyal, P., Girshick, R., He, K., Dollar, P. (2020). Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 42 (2), 318–327. doi: 10.1109/TPAMI.2018.2858826
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C. Y., et al. (2016). “SSD: Single shot multibox detector,” in Lecture notes in computer science (Including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) , vol. 9905 LNCS. (Amsterdam, Netherlands: Springer), 21–37. doi: 10.1007/978-3-319-46448-0_2
Liu, B., Ding, Z., Tian, L., He, D., Li, S., Wang, H. (2020). Grape leaf disease identification using improved deep convolutional neural networks. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01082
Liu, J., Wang, X. (2020). Tomato diseases and pests detection based on improved yolo V3 convolutional neural network. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00898
Liu, J., Wang, X. (2021a). Early recognition of tomato gray leaf spot disease based on MobileNetv2 − YOLOv3 model. Plant Methods 2020, 1–16. doi: 10.1186/s13007-020-00624-2
Liu, J., Wang, X. (2021b). Plant diseases and pests detection based on deep learning: a review. Plant Methods 17 (1), 1–18. doi: 10.1186/s13007-021-00722-9
Liu, W., Wang, Z., Liu, X., Zeng, N., Liu, Y., Alsaadi, F. E. (2017). Neurocomputing a survey of deep neural network architectures and their applications ☆. Neurocomputing 234 (November 2016), 11–26. doi: 10.1016/j.neucom.2016.12.038
Martinelli, F., Scalenghe, R., Davino, S., Panno, S., Scuderi, G., Ruisi, P., et al. (2015). Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 35, 1–25. doi: 10.1007/s13593-014-0246-1
Masilamani, G. K., Valli, R. (2021). “Art classification with pytorch using transfer learning,” in 2021 International Conference on System, Computation, Automation and Networking (ICSCAN). (Puducherry, India: IEEE), 1–5. doi: 10.1109/ICSCAN53069.2021.9526457
Mohanty, S. P., Hughes, D. P., Salathé, M. (2016). Using deep learning for image-based plant disease detection. Front. Plant Sci. 7 (September). doi: 10.3389/fpls.2016.01419
Nasirahmadi, A., Wilczek, U., Hensel, O. (2021). Sugar beet damage detection during harvesting using different convolutional neural network models. Agric. (Switzerland) 11 (11), 1–13. doi: 10.3390/agriculture11111111
Navab, N., Hornegger, J., Wells, W. M., Frangi, A. F. (2015) in 18th International Conference, Munich, Germany, October 5-9, 2015, (Germany: Springer) 9351, 12–20, proceedings, part III. doi: 10.1007/978-3-319-24574-4
Oppenheim, D., Shani, G., Erlich, O., Tsror, L. (2019). Using deep learning for image-based potato tuber disease detection. Phytopathology 109 (6), 1083–1087. doi: 10.1094/PHYTO-08-18-0288-R
Peng, Y., Wang, Y. (2022). Leaf disease image retrieval with object detection and deep metric learning. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.963302
Permanasari, Y., Ruchjana, B. N., Hadi, S., Rejito, J. (2022). Innovative region convolutional neural network algorithm for object identification. J. Open Innovation: Technology Market Complexity 8 (4), 182. doi: 10.3390/joitmc8040182
Picon, A., Seitz, M., Alvarez-gila, A., Mohnke, P., Ortiz-barredo, A., Echazarra, J. (2019). Crop conditional convolutional neural networks for massive multi-crop plant disease classi fi cation over cell phone acquired images taken on real fi eld conditions. Comput. Electron. Agric. 167, 105093. doi: 10.1016/j.compag.2019.105093
Pu, Y., Gan, Z., Henao, R., Yuan, X., Li, C., Stevens, A., et al. (2016). Variational autoencoder for deep learning of images, labels and captions. Adv. Neural Inf. Process. Syst. 29, 1–13. doi: 10.48550/arXiv.1609.08976
Sabrol, H. (2015). Recent studies of image and soft computing techniques for plant disease recognition and classification. Int. J. Comput. Appl. 126, 1, 44–55. doi: 10.5120/ijca2015905982
Salakhutdinov, R., Larochelle, H. (2010). “Efficient learning of deep Boltzmann machines,” in Proceedings of the thirteenth international conference on artificial intelligence and statistics, Sardinia, Italy: mlr press. 1–40.
Sarker, I. H. (2021). Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2 (6), 1–20. doi: 10.1007/s42979-021-00815-1
Shelhamer, E., Long, J., Darrell, T. (2017). Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 39 (4), 640–651. doi: 10.48550/arXiv.1411.4038
Shoaib, M., Hussain, T., Shah, B., Ullah, I., Shah, S. M., Ali, F., et al. (2022a). Deep learning-based segmentation and classification of leaf images for detection of tomato plant disease. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1031748
Shoaib, M., Sayed, N. (2022). Traitement du signal YOLO object detector and inception-V3 convolutional neural network for improved brain tumor segmentation. Traitement Du Signal 39, 1, 371–380. doi: 10.18280/ts.390139
Shoaib, M., Shah, B., Hussain, T., Ali, A., Ullah, A., Alenezi, F., et al. (2022b). A deep learning-based model for plant lesion segmentation , subtype identi fi cation , and survival probability estimation 1–15. doi: 10.3389/fpls.2022.1095547
Shougang, R., Fuwei, J., Xingjian, G., Peishen, Y., Wei, X., Huanliang, X. (2020). Deconvolution-guided tomato leaf disease identification and lesion segmentation model. J. Agric. Eng. 36 (12), 186–195. doi: 10.1186/s13007-021-00722-9
Siddiqua, A., Kabir, M. A., Ferdous, T., Ali, I. B., Weston, L. A. (2022). Evaluating plant disease detection mobile applications: Quality and limitations. Agronomy 12 (8), 1869. doi: 10.3390/agronomy12081869
Singh, D., Jain, N., Jain, P., Kayal, P., Kumawat, S., Batra, N. (2020). “PlantDoc: A dataset for visual plant disease detection,” in Proceedings of the 7th ACM IKDD CoDS and 25th COMAD. (Hyderabad, India: Association for computing Machinery) 249–253.
Singh, V., Misra, A. K. (2017). Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 4 (1), 41–49. doi: 10.1016/j.inpa.2016.10.005
Sladojevic, S., Arsenovic, M., Anderla, A., Culibrk, D., Stefanovic, D. (2016). Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 1–12. doi: 10.1155/2016/3289801
Soliman, M. M., Kamal, M. H., Nashed, M. A. E. M., Mostafa, Y. M., Chawky, B. S., Khattab, D. (2019). “Violence recognition from videos using deep learning techniques,” in 2019 Ninth International Conference on Intelligent Computing and Information Systems (ICICIS), (Cairo, Egypt: IEEE). 80–85. doi: 10.1109/ICICIS46948.2019.9014714
Stančić, A., Vyroubal, V., Slijepčević, V. (2022). Classification efficiency of pre-trained deep CNN models on camera trap images. J. Imaging 8 (2), 20. doi: 110.3390/jimaging8020020
Stewart, E. L., Wiesner-Hanks, T., Kaczmar, N., DeChant, C., Wu, H., Lipson, H., et al. (2019). Quantitative phenotyping of northern leaf blight in UAV images using deep learning. Remote Sens. 11 (19), 2209. doi: 10.3390/rs11192209
Szymak, P., Piskur, P., Naus, K. (2020). The effectiveness of using a pretrained deep learning neural networks for object classification in underwater video. Remote Sens. 12 (18), 1–19. doi: 10.3390/RS12183020
Tahir, A. M., Qiblawey, Y., Khandakar, A., Rahman, T., Khurshid, U., Musharavati, F., et al. (2022). Deep learning for reliable classification of COVID-19, MERS, and SARS from chest X-ray images. Cogn. Comput. 2019, 17521772. doi: 10.1007/s12559-021-09955-1
Tianjiao, C., Wei, D., Juan, Z., Chengjun, X., Rujing, W., Wancai, L. (2019). Intelligent identification system of disease and insect pests based on deep learning. China Plant Prot Guide 39 (004), 26–34.
Too, E. C., Yujian, L., Njuki, S., Yingchun, L. (2019). A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 161, 272–279. doi: 10.1016/j.compag.2018.03.032
Tran, D., Bourdev, L., Fergus, R., Torresani, L., Paluri, M. (2015). “Learning spatiotemporal features with 3d convolutional networks,” in Proceedings of the IEEE international conference on computer vision. 4489–4497.
Ullah, A., Muhammad, K., Haq, I. U., Baik, S. W. (2019). Action recognition using optimized deep autoencoder and CNN for surveillance data streams of non-stationary environments. Future Generation Comput. Syst. 96, 386–397. doi: 10.1016/j.future.2019.01.029
Wang, B. (2022). Identification of crop diseases and insect pests based on deep learning. Sci. Program. 2022, 1–10. doi: 10.1155/2022/9179998
Wang, Q., Qi, F., Sun, M., Qu, J., Xue, J. (2019). Identification of tomato disease types and detection of infected areas based on deep convolutional neural networks and object detection techniques. Comput. Intell. Neurosci. 2019, 9142753. doi: 10.1155/2019/9142753
Wang, G., Sun, Y., Wang, J. (2017). Automatic image-based plant disease severity estimation using deep learning. Comput. Intell. Neurosci. 2017, 1–9. doi: 10.1155/2017/2917536
Wang, L., Xiong, Y., Wang, Z., Qiao, Y. (2015). Towards good practices for very deep two-stream ConvNets. arXiv preprint arXiv:1507.02159. , 1–5. doi: 10.48550/arXiv.1507.02159
Wang, Z., Zhang, S. (2018). Segmentation of corn leaf disease based on fully convolution neural network. Acad. J. Comput. Inf Sci. 1, 9–18. doi: 10.25236/AJCIS.010002
Wiesner-hanks, T., Wu, H., Stewart, E., Dechant, C., Kaczmar, N., Lipson, H., et al. (2019). Millimeter-level plant disease detection from aerial photographs via deep learning and crowdsourced data. Front. Plant Sci. 10, 1–11. doi: 10.3389/fpls.2019.01550
Xie, X., Ma, Y., Liu, B., He, J., Li, S., Wang, H. (2020). A deep-Learning-Based real-time detector for grape leaf diseases using improved convolutional neural networks. Front. Plant Sci. 11, 1–14. doi: 10.3389/fpls.2020.00751
Xu, J., Chen, J., You, S., Xiao, Z., Yang, Y., Lu, J. (2021). Robustness of deep learning models on graphs: A survey. AI Open 2 (December 2020), 69–78. doi: 10.1016/j.aiopen.2021.05.002
Yalcin, H., Razavi, S. (2016). “Plant classification using convolutional neural networks,” in 2016 Fifth International Conference on Agro-Geoinformatics (Agro-Geoinformatics). (Tianjin, China: IEEE), 1–5. doi: 10.1109/Agro-Geoinformatics.2016.7577698
Yoosefzadeh-Najafabadi, M., Earl, H. J., Tulpan, D., Sulik, J., Eskandari, M. (2021). Application of machine learning algorithms in plant breeding: Predicting yield from hyperspectral reflectance in soybean. Front. Plant Sci. 11 (January). doi: 10.3389/fpls.2020.624273
Zhang, S., Huang, W., Zhang, C. (2019). ScienceDirect three-channel convolutional neural networks for vegetable leaf disease recognition. Cogn. Syst. Res. 53, 31–41. doi: 10.1016/j.cogsys.2018.04.006
Zhao, S., Wu, Y., Ede, J. M., Balafrej, I., Alibart, F. (2021). Research on image classification algorithm based on pytorch research on image classification algorithm based on pytorch. Journal of Physics: Conference Series, 4th International Conference on Computer Information Science and Application Technology (CISAT 2021). 2010, 1–6. doi: 10.1088/1742-6596/2010/1/012009
Zhou, G., Zhang, W., Chen, A., He, M., Ma, X. (2019). Rapid detection of rice disease based on FCM-KM and faster r-CNN fusion. IEEE Access 7, 143190–143206. doi: 10.1109/ACCESS.2019.2943454
Keywords: machine learning, deep learning, plant disease detection, image processing, convolutional neural networks, performance evaluation, practical applications
Citation: Shoaib M, Shah B, EI-Sappagh S, Ali A, Ullah A, Alenezi F, Gechev T, Hussain T and Ali F (2023) An advanced deep learning models-based plant disease detection: A review of recent research. Front. Plant Sci. 14:1158933. doi: 10.3389/fpls.2023.1158933
Received: 04 February 2023; Accepted: 27 February 2023; Published: 21 March 2023.
Reviewed by:
Copyright © 2023 Shoaib, Shah, EI-Sappagh, Ali, Ullah, Alenezi, Gechev, Hussain and Ali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Farman Ali, [email protected] ; Tariq Hussain, [email protected]
† These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Pepper leaf disease recognition based on enhanced lightweight convolutional neural networks
Affiliations.
- 1 College of Mechanical Engineering, Yangzhou University, Yangzhou, China.
- 2 Nanjing Institute of Agricultural Mechanization, Ministry of Agriculture and Rural Affairs, Nanjing, China.
- 3 Faculty of Science and Engineering, Manchester Metropolitan University Manchester, Manchester, United Kingdom.
- PMID: 37621882
- PMCID: PMC10445539
- DOI: 10.3389/fpls.2023.1230886
Pepper leaf disease identification based on convolutional neural networks (CNNs) is one of the interesting research areas. However, most existing CNN-based pepper leaf disease detection models are suboptimal in terms of accuracy and computing performance. In particular, it is challenging to apply CNNs on embedded portable devices due to a large amount of computation and memory consumption for leaf disease recognition in large fields. Therefore, this paper introduces an enhanced lightweight model based on GoogLeNet architecture. The initial step involves compressing the Inception structure to reduce model parameters, leading to a remarkable enhancement in recognition speed. Furthermore, the network incorporates the spatial pyramid pooling structure to seamlessly integrate local and global features. Subsequently, the proposed improved model has been trained on the real dataset of 9183 images, containing 6 types of pepper diseases. The cross-validation results show that the model accuracy is 97.87%, which is 6% higher than that of GoogLeNet based on Inception-V1 and Inception-V3. The memory requirement of the model is only 10.3 MB, which is reduced by 52.31%-86.69%, comparing to GoogLeNet. We have also compared the model with the existing CNN-based models including AlexNet, ResNet-50 and MobileNet-V2. The result shows that the average inference time of the proposed model decreases by 61.49%, 41.78% and 23.81%, respectively. The results show that the proposed enhanced model can significantly improve performance in terms of accuracy and computing efficiency, which has potential to improve productivity in the pepper farming industry.
Keywords: GoogLeNet; crop disease recognition; deep convolutional neural networks; lightweight neural networks; real-time recognition.
Copyright © 2023 Dai, Sun, Wang, Dorjoy, Zhang, Miao, Han, Zhang and Wang.
PubMed Disclaimer
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The overview process of the…
The overview process of the improved model.
Related images of pepper leaf…
Related images of pepper leaf disease. (A) Pepper scab, (B) Pepper powdery mildew,…
Image enhancement. (A) Original image,…
Image enhancement. (A) Original image, (B) 90-degree rotation, (C) Random crop, (D) Color…
GoogLeNet structure.
GoogLeNet-EL structure.
The structures of the Inception…
The structures of the Inception module and optimized Inception module. (A) a classical…
Cropping of pepper leaf disease…
Cropping of pepper leaf disease image.
The network structure with SPP.
Activation function. (A) ReLu (B)…
Activation function. (A) ReLu (B) LeakyRelu.
A comparison graph of the…
A comparison graph of the training results of the GoogLeNet-EL, Inception-V1, and Inception-V3.
A comparison graph of the necessity test of GoogLeNet-EL related improvement aspects.
Comparison of test results with…
Comparison of test results with other models.
Confusion matrix for different models…
Confusion matrix for different models on test set. (a) Pepper scab, (b) Pepper…
Similar articles
- Early stage black pepper leaf disease prediction based on transfer learning using ConvNets. Kini AS, Prema KV, Pai SN. Kini AS, et al. Sci Rep. 2024 Jan 16;14(1):1404. doi: 10.1038/s41598-024-51884-0. Sci Rep. 2024. PMID: 38228767 Free PMC article.
- Convolutional neural network in rice disease recognition: accuracy, speed and lightweight. Ning H, Liu S, Zhu Q, Zhou T. Ning H, et al. Front Plant Sci. 2023 Nov 1;14:1269371. doi: 10.3389/fpls.2023.1269371. eCollection 2023. Front Plant Sci. 2023. PMID: 38023901 Free PMC article. Review.
- Holographic Microwave Image Classification Using a Convolutional Neural Network. Wang L. Wang L. Micromachines (Basel). 2022 Nov 23;13(12):2049. doi: 10.3390/mi13122049. Micromachines (Basel). 2022. PMID: 36557348 Free PMC article.
- Classification and Visualisation of Normal and Abnormal Radiographs; A Comparison between Eleven Convolutional Neural Network Architectures. Ananda A, Ngan KH, Karabağ C, Ter-Sarkisov A, Alonso E, Reyes-Aldasoro CC. Ananda A, et al. Sensors (Basel). 2021 Aug 9;21(16):5381. doi: 10.3390/s21165381. Sensors (Basel). 2021. PMID: 34450821 Free PMC article.
- Grape Leaf Disease Identification Using Improved Deep Convolutional Neural Networks. Liu B, Ding Z, Tian L, He D, Li S, Wang H. Liu B, et al. Front Plant Sci. 2020 Jul 15;11:1082. doi: 10.3389/fpls.2020.01082. eCollection 2020. Front Plant Sci. 2020. PMID: 32760419 Free PMC article.
- Identification of wheat seedling varieties based on MssiapNet. Feng Y, Liu C, Han J, Lu Q, Xing X. Feng Y, et al. Front Plant Sci. 2024 Jan 18;14:1335194. doi: 10.3389/fpls.2023.1335194. eCollection 2023. Front Plant Sci. 2024. PMID: 38304454 Free PMC article.
- Abade A., Ferreira P. A., Vidal F. (2021). Plant diseases recognition on images using convolutional neural networks: A systematic review. Comput. Electron. Agr. 185, 106125. doi: 10.1016/j.compag.2021.106125 - DOI
- Ak A., Kbw A., Zw A., Mc B. (2019). Deep learning – Method overview and review of use for fruit detection and yield estimation. Comput. Electron. Agr. 162, 219–234. doi: 10.1016/j.compag.2019.04.017 - DOI
- Bera S., Shrivastava V. K. (2020). Analysis of various optimizers on deep convolutional neural network model in the application of hyperspectral remote sensing image classification. Int. J. Remote Sens. 41, 2664–2683. doi: 10.1080/01431161.2019.1694725 - DOI
- Bhujel A., Kim N. E., Arulmozhi E., Basak J. K., Kim H. T. (2022). A lightweight attention-based convolutional neural networks for tomato leaf disease classification. Agriculture-basel 12 (2), 228. doi: 10.3390/agriculture12020228 - DOI
- Chang W.-Y., Wu S.-J., Hsu J.-W. (2020). Investigated iterative convergences of neural network for prediction turning tool wear. Int. J. Advanced Manufacturing Technol. 106, 2939–2948. doi: 10.1007/s00170-019-04821-9 - DOI
Related information
Grants and funding, linkout - more resources, full text sources.
- Europe PubMed Central
- Frontiers Media SA
- PubMed Central
Miscellaneous
- NCI CPTAC Assay Portal

- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Plants (Basel)

Plant Disease Detection and Classification by Deep Learning
Muhammad hammad saleem.
1 Department of Mechanical and Electrical Engineering, School of Food and Advanced Technology, Massey University, Auckland 0632, New Zealand; [email protected]
Johan Potgieter
2 Massey Agritech Partnership Research Centre, School of Food and Advanced Technology, Massey University, Palmerston North 4442, New Zealand; [email protected]

Khalid Mahmood Arif
Plant diseases affect the growth of their respective species, therefore their early identification is very important. Many Machine Learning (ML) models have been employed for the detection and classification of plant diseases but, after the advancements in a subset of ML, that is, Deep Learning (DL), this area of research appears to have great potential in terms of increased accuracy. Many developed/modified DL architectures are implemented along with several visualization techniques to detect and classify the symptoms of plant diseases. Moreover, several performance metrics are used for the evaluation of these architectures/techniques. This review provides a comprehensive explanation of DL models used to visualize various plant diseases. In addition, some research gaps are identified from which to obtain greater transparency for detecting diseases in plants, even before their symptoms appear clearly.
1. Introduction
The Deep Learning (DL) approach is a subcategory of Machine Learning (ML), introduced in 1943 [ 1 ] when threshold logic was introduced to build a computer model closely resembling the biological pathways of humans. This field of research is still evolving; its evolution can be divided into two time periods-from 1943–2006 and from 2012–until now. During the first phase, several developments like backpropagation [ 2 , 3 ], chain rule [ 4 ], Neocognitron [ 5 ], hand written text recognition (LeNET architecture) [ 6 ], and resolving the training problem [ 7 , 8 ] were observed (as shown in Figure 1 ). However, in the second phase, state-of-the-art algorithms/architectures were developed for many applications including self-driving cars [ 9 , 10 , 11 ], healthcare sector [ 12 , 13 , 14 ], text recognition [ 6 , 15 , 16 , 17 ], earthquake predictions [ 18 , 19 , 20 ], marketing [ 21 ], finance [ 22 , 23 ], and image recognition [ 24 , 25 , 26 , 27 , 28 , 29 ]. Among those architectures, AlexNet [ 30 ] is considered to be a breakthrough in the field of DL as it won the ImageNet challenge for object recognition known as ImageNet Large Scale Visual Recognition Challenge (ILSVRC) in the year 2012. Soon after, several architectures were introduced to overcome the loopholes observed previously. For the evaluation of these algorithms/architectures, various performance metrics were used. Among these metrics, top-1%/top-5% error [ 24 , 26 , 30 , 31 ], precision and recall [ 25 , 32 , 33 , 34 ], F1 score [ 32 , 35 ], training/validation accuracy and loss [ 34 , 36 ], classification accuracy (CA) [ 37 , 38 , 39 , 40 , 41 ] are the most popular. For the implementation of DL models, several steps are required, from the collection of datasets to visualization mappings are explained in Figure 2 .

Summary of the evolution of deep learning from 1943–2006.

Flow diagram of DL implementation: First, the dataset is collected [ 25 ] then split into two parts, normally into 80% of training and 20% of validation set. After that, DL models are trained from scratch or by using transfer learning technique, and their training/validation plots are obtained to indicate the significance of the models. Then, performance metrics are used for the classification of images (type of particular plant disease), and finally, visualization techniques/mappings [ 55 ] are used to detect/localize/classify the images.
When DL architectures started to evolve with the passage of time, researchers applied them to image recognition and classification. These architectures have also been implemented for different agricultural applications. For example, in [ 42 ], classification of leaves was performed by using author-modified CNN and Random Forest (RF) classifier among 32 species in which the performance was evaluated through CA at 97.3%. On the other hand, it was not as efficient at detecting occluded objects [ 43 ]. Leaf and fruit counting were also performed by deep CNN in [ 44 , 45 ] and [ 46 ] respectively. For classification of crop type, [ 47 ] used author-modified CNN, [ 36 ] applied VGG 16, [ 34 ] implemented three unit LSTM, and [ 33 ] used CNN and RGB histogram technique. [ 47 ] used CA, [ 36 ] used CA and Intersection over Union (IoU), [ 34 ] used CA and F1, and [ 33 ] used F1-score as a performance metric. Among them, [ 33 , 47 ] did not provide training/validation accuracy and loss. Moreover, recognition of different plants has been done by the DL approach in [ 48 , 49 , 50 ]. [ 48 , 50 ] employed user-modified CNN while [ 49 ] used AlexNet architecture. All were evaluated on the basis of CA. [ 49 ] outperformed the other two in terms of CA. Similarly, crop/weed discrimination was performed in [ 51 , 52 ], in which the author proposed CNN be used, and two datasets were utilized for the evaluation of the model. [ 51 ] evaluated precision and recall; however, [ 52 ] obtained CA for the validation of the proposed models respectively. The identification of plants by the DL approach was studied and achieved a success rate of 91.78% [ 53 ]. On top of that, DL approaches are also used for critical tasks like plant disease detection and classification, which is the main focus of this review. There are some research papers previously presented to summarize the research based on agriculture (including plant disease recognition) by DL [ 43 , 54 ], but they lacked some of the recent developments in terms of visualization techniques implemented along with the DL and modified/cascaded version of famous DL models, which were used for plant disease identification. Moreover, this review also provides the research gaps in order to get a clearer/more transparent vision of symptoms observed due to diseases in the plants.
The remaining part of the paper is comprised of Section 2 , describing the famous and new/modified DL architectures along with visualization mapping/techniques used for plant disease detection; Section 3 , elaborating upon the Hyperspectral Imaging with DL models; and finally, Section 4 , concluding the review and providing future recommendations for achieving more advancements in the visualization, detection, and classification of plants’ diseases.
2. Plant Disease Detection by Well-Known DL Architectures
Many state-of-the-art DL models/architectures evolved after the introduction of AlexNet [ 30 ] (as shown in Figure 3 and Table 1 ) for image detection, segmentation, and classification. This section presents the researches done by using famous DL architectures for the identification and classification of plants’ diseases. Moreover, there are some related works in which new visualization techniques and modified/improved versions of DL architectures were introduced to achieve better results. Among all of them, the PlantVillage dataset has been used widely as it contains 54,306 images of 14 different crops having 26 plant diseases [ 25 ]. Moreover, they used several performance metrics to evaluate the selected DL models, which are described as below.

Summary of the evolution of various deep learning models from 2012 until now.
Comparison of state-of-the-art deep learning models.
| Deep Learning Models | Parameters | Key Features and Pros/Cons |
|---|---|---|
| LeNet | 60k | First CNN model. Few parameters as compared to other CNNmodels. Limited capability of computation |
| AlexNet | 60M | Known as the first modern CNN. Best image recognition performance at its time. Used ReLU to achieve better performance. Dropout technique was used to avoid overfitting |
| OverFeat | 145M | First model used for detection, localization, and classification of objects through a single CNN. Large number of parameters as compared to AlexNet |
| ZFNet | 42.6M | Reduced weights (as compared to AlexNet) by considering 7 × 7 kernels and improved accuracy |
| VGG | 133M–144M | 3 × 3 receptive fields were considered to include more number of non-linearity functions which made decision function discriminative. Computationally expensive model due to large number of parameters |
| GoogLeNet | 7M | Fewer number of parameters as compared to AlexNet model. Better accuracy at its time |
| ResNet | 25.5M | Vanishing gradient problem was addressed. Better accuracy than VGG and GoogLeNet models |
| DenseNet | 7.1M | Dense connections between the layers. Reduced number of parameters with better accuracy |
| SqueezeNet | 1.25M | Similar accuracy as AlexNet with 50 times lesser parameters. Considered 1 × 1 filters instead of 3 × 3 filters. Input channels were decreased. Large activation maps of convolution layers |
| Xception | 22.8M | A depth-wise separable convolution approach. Performed better than VGG, ResNet, and Inception-v3 models |
| MobileNet | 4.2M | Considered the depth-wise separable convolution concept. Reduced parameters significantly. Achieved accuracy near to VGG and GoogLeNet |
| Modified/Reduced MobileNet | 0.5/0.54M | Lesser number of parameters as compared to MobileNet. Similar accuracy as compared to MobileNet |
| VGG-Inception | 132M | A cascaded version of VGG and inception module. The number of parameters were reduced by substituting 5 × 5 convolution layers with two 3 × 3 layers. Testing accuracy was increased as compared to many well-known DL models like AlexNet, GoogLeNet, Inception-v3, ResNet, and VGG-16. |
2.1. Implementation of DL Models
2.1.1. without visualization technique.
In [ 56 ], CNN was used for the classification of diseases in maize plants and histogram techniques to show the significance of the model. In [ 57 ], basic CNN architectures like AlexNet, GoogLeNet and ResNet were implemented for identifying the tomato leaf diseases. Training/validation accuracy were plotted to show the performance of the model; ResNet was considered as the best among all the CNN architectures. In order to detect the diseases in banana leaf, LeNet architecture was implemented and CA, F1-score were used for the evaluation of the model in Color and Gray Scale modes [ 32 ]. Five CNN architectures were used in [ 58 ], namely, AlexNet, AlexNetOWTbn, GoogLeNet, Overfeat, and VGG architectures in which VGG outclassed all the other models. In [ 35 ], eight different plant diseases were recognized by three classifiers, Support Vector Machines (SVM), Extreme Learning Machine (ELM), and K-Nearest Neighbor (KNN)), used with the state-of-the-art DL models like GoogLeNet, ResNet-50, ResNet-101, Inception-v3, InceptionResNetv2, and SqueezeNet. A comparison was made between those models, and ResNet-50 with SVM classifier got the best results in terms of performance metrics like sensitivity, specificity, and F1-score. According to [ 59 ], a new DL model—Inception-v3—was used for the detection of cassava disease. In [ 60 ], plant diseases in cucumber were classified by the two basic versions of CNN and got the highest accuracy, equal to 0.823. The traditional plant disease recognition and classification method was replaced by Super-Resolution Convolutional Neural Network (SRCNN) in [ 61 ]. For the classification of tomato plant disease, AlexNet and SqueezeNet v1.1 models were used in which AlexNet was found to be the better DL model in terms of accuracy [ 62 ]. A comparative analysis was presented in [ 63 ] to select the best DL architecture for detection of plant diseases. Moreover in [ 64 ], six tomato plant diseases were classified by using AlexNet and VGG-16 DL architectures, and a detailed comparison was provided with the help of classification accuracy. In the above approaches, no visualization technique was applied to spot the symptoms of diseases in the plants.
2.1.2. With Visualization Techniques
The following approaches employed DL models/architectures and also visualization techniques which were introduced for a clearer understanding of plants’ diseases. For example, [ 55 ] introduced the saliency map for visualizing the symptoms of plant disease; [ 27 ] identified 13 different types of plant disease with the help of CaffeNet CNN architecture, and achieved CA equal to 96.30%, which was better than the previous approach like SVM. Moreover, several filters were used to indicate the disease spots. Similarly, [ 25 ] used AlexNet and GoogLeNet CNN architectures by using the publicly available PlantVillage dataset. The performance was evaluated by means of precision (P), recall (R), F1 score, and overall accuracy. The uniqueness of this paper was the implication of three scenarios (color, grayscale, and segmented) for evaluating the performance metrics and comparison of the two famous CNN architectures. It was concluded that GoogLeNet outperformed AlexNet. Moreover, visualization activation in the first layers clearly showed the spots of diseases. In [ 65 ], a modified LeNet model was used to detect olive plant diseases. The segmentation and edges maps were used to spot the diseases in the plants. Detection of four cucumber diseases was done in [ 66 ] and accuracy was compared with Random Forest, Support Vector Machines, and AlexNet models. Moreover, the image segmentation method was used to view the symptoms of diseases in the plants. A new DL model was introduced in [ 67 ] named teacher/student network and proposed a novel visualization method to identify the spots of plant diseases. DL models with some detectors were implemented in [ 68 ], in which the diseases in plants were marked along with their prediction percentage. Three detectors, named Faster-RCNN, RFCN and SSD, were used with the famous architectures like AlexNet, GoogLeNet, VGG, ZFNet, ResNet-50, ResNet-101 and ResNetXt-101 for a comparative study which outlined the best among all the selected architectures. It was concluded that ResNet-50 with the detector R-FCN gave the best results. Furthermore, a kind of bounding box was drawn to identify the particular type of disease in the plants. In [ 69 ], a banana leaf disease and pest detection was performed by using three CNN models (ResNet-50, Inception-V2 and MobileNet-V1) with Faster-RCNN and SSD detectors. According to [ 70 ], different combinations of CNN were used and presented heat maps as input to the diseased plants’ images and provided the probability related to the occurrence of a particular type of disease. Moreover, ROC curve evaluates the performance of the model. Furthermore, feature maps for rice disease were also included in the paper. LeNet model was used in [ 71 ] to detect and classify diseases in the soybean plant. In [ 72 ], a comparison between AlexNet and GoogLeNet architectures for tomato plant diseases was done, in which GoogLeNet performed better than the AlexNet; also, it proposed occlusion techniques to recognize the regions of diseases. The VGG-FCN and VGG-CNN models were implemented in [ 73 ], for the detection of wheat plant diseases and visualization of features in each block. In [ 74 ], VGG-CNN model was used for the detection of Fusarium wilt in radish and K-means clustering method was used to show the marks of diseases. A semantic segmentation approach by CNN was proposed in [ 75 ] to detect the disease in cucumber. In [ 76 ], an approach based on the individual symptoms/spots of diseases in the plants was introduced by using a DL model for detecting plant diseases. A Deep CNN framework was developed for identification, classification, and quantification of eight soybean stresses in [ 77 ]. In [ 78 ], rice plant diseases were identified by CNN, and feature maps were obtained to identify the patches of diseases. A deep residual neural network was extended in [ 79 ] for the development of a mobile application in which a clear identification of diseases in plants was done by the hot spot. An algorithm based on the hot spot technique was also used in [ 80 ], in which those spots were extracted by modification in the segmented image to attain color constancy. Furthermore, each obtained hot-spot was described by two descriptors, one was used to evaluate the color information of the disease and other was used to identify the texture of the hot-spots. The cucumber plant diseases were identified in [ 81 ] by using the dilation convolutional neural network. A state-of-the-art visualization technique was proposed in [ 82 ] by correlation coefficient and DL models like AlexNet and VGG-16 architectures. In [ 83 ], color space and various vegetation indices combined with CNN model (LeNet) to detect the diseases in grapes. To summarize, Table 2 outlines some of the visualization mapping/techniques.
Visualization mapping/techniques used in several approaches.
| Visualization Techniques/Mappings | References |
|---|---|
| Visualization of features having filter from first to final layer | [ ] |
| Visualize activations in first convolutional layer | [ ] |
| Saliency map visualization | [ ] |
| Classification and localization of diseases by bounding boxes | [ ] |
| Heat maps were used to identify the spots of the disease | [ ] |
| Feature map for the diseased rice plant | [ ] |
| Symptoms visualization method | [ ] |
| Feature and spatial core maps | [ ] |
| Color space into HSV and K-means clustering | [ ] |
| Feature map for spotting the diseases | [ ] |
| Image segmentation method | [ ] |
| Reconstruction of images on discriminant regions, segmentation of images by binary threshold theorem, and heat map construction | [ ] |
| Saliency map visualization | [ ] |
| Saliency map, 2D and 3D contour, mesh graph image | [ ] |
| Activation visualization | [ ] |
| Segmentation map and edge map | [ ] |
For the practical experimentation of detection of plants’ diseases, an actual/real background/environment should be considered in order to evaluate the performance of the DL model more accurately. In most of the above approaches, the selected datasets considered plain backgrounds which are not realistic scenarios for identification and classification of the diseases [ 25 , 27 , 32 , 56 , 57 , 58 , 60 , 61 , 65 , 72 , 77 , 78 ], except for a few of them that have considered the original backgrounds [ 35 , 59 , 68 , 70 , 73 , 74 ]. The output of the visualization techniques used in several researches are shown in Figure 4 , Figure 5 , Figure 6 , Figure 7 , Figure 8 , Figure 9 , Figure 10 and Figure 11 .

Feature maps after the application of convolution to an image: ( a ) real image, ( b ) first convolutional layer filter, ( c ) rectified output from first layer, ( d ) second convolutional layer filter, ( e ) output from second layer, ( f ) output of third layer, ( g ) output of fourth layer, ( h ) output of fifth layer [ 27 ].

Tomato plant disease detection by heat map: on left hand side ( a ) tomato early blight, ( b ) tomato septoria leaf spot, ( c ) tomato late blight and ( d ) tomato leaf mold) and saliency map; on right hand side ( a ) tomato healthy, ( b ) tomato late blight, ( c ) tomato early blight, ( d ) tomato septoria leaf spot, ( e ) tomato early blight, ( f ) tomato leaf mold) [ 55 ].

Detection of maize disease (indicated by red circles) by heat map [ 70 ].

Bounding box indicates the type of diseases along with the probability of their occurrence [ 68 ]. A bounding box technique was used in Figure 7 in which ( a ) represents the one type of disease along with its rate of occurrence, ( b ) indicates three types of plant disease (miner, temperature, and gray mold) in a single image, ( c , d ) shows one class of disease but contains different patterns on the front and back side of the image, ( e , f ) displays different patterns of gray mold in the starting and end stages [ 68 ].

( a ) Teacher/student architecture approach; ( b ) segmentation using a binary threshold algorithm [ 67 ].

Comparison of Teacher/student approach visualization map with the previous approaches [ 67 ].

Activation visualization for detection of apple plant disease to show the significance of a VGG-Inception model (the plant disease is indicated by the red circle) [ 85 ].

Segmentation and edge map for olive leaf disease detection [ 65 ].
In Figure 4 , feature maps from the first to the fifth hidden layer are shown as the neuron in a feature map having identical features at different positions of an image. Starting from the first layer (a), the features in feature maps represent separate pixels to normal lines, whereas the fifth layer shows some particular parts of the image (h).
Two types of visualization maps are shown in Figure 5 , namely, heat map and saliency map techniques. The heat maps identify the diseases shown as red boxes in the input image, but it should be noted that one disease marked in (d) has not been detected. This problem was resolved in the saliency map technique after the application of the guided back-propagation [ 55 ]; all the spots of plant disease were successfully identified thanks to a method which is superior to the heat map.
Figure 6 represents the heat map to detect the disease in maize plants. First, the image was represented in the form of the probability of each portion containing disease. Then, the probabilities were placed into the form of a matrix in order to denote the outcome of all the areas of the input image.
A new visualization technique was proposed in [ 67 ] as shown in Figure 8 and Figure 9 . In Figure 8 a, the input image was regenerated for student/teacher architecture [ 67 ], and a single channel heat map was produced after the application of simple aggregation on the channels of the regenerated image ( Figure 8 b). Then, a simple binary threshold algorithm was applied to obtain sharp symptoms of diseases in the plant. Then, [ 67 ] indicated the significance of the proposed technique by comparing it with the other visualization techniques as shown in Figure 9 . On the left hand side, LRP-Z, LRP-Epsilon, and gradient did not identify plant diseases clearly. However, the Deep Taylor approach produced better results but indicated some portion of the leaf disease. On the right hand side, an imperfect localization of the plant disease was shown in grad-cam techniques which was resolved in the proposed technique by the use of a decoder [ 67 ].
In order to find the significance of CNN architectures to differentiate between various diseases of plants, the feature maps were obtained as shown in Figure 10 . The result proves a good performance of the proposed CNN model as it clearly identifies the disease in plants [ 85 ].
In Figure 11 the segmentation and edged maps were obtained to identify the diseases in plants. It is noted that the yellow colored area is marked as white surface in the segmentation map to show the affected part of the leaf.
2.2. New/Modified DL Architectures for Plant-Disease Detection
According to some of the research papers, new/modified DL architectures have been introduced to obtain better/transparent detection of plant disease, such as [ 86 ] presented improved GoogLeNet and Cifar-10 models and their performance compared with AlexNet and VGG. It was found that improved versions of these state-of-the-art models produced a remarkable accuracy of 98.9%. In [ 87 ], a new DL model was introduced to obtain more accurate detection of plant diseases as compared to SVM, AlexNet, GoogLeNet, ResNet-20, and VGG-16 models. This model achieved 97.62% accuracy for classifying apple plant diseases. Moreover, the dataset extended in 13 different ways (rotation of 90°, 180°, 270° and mirror symmetry (horizontal symmetry), change in contrast, sharpness and brightness). Moreover, the whole dataset was transformed into Gaussian noise and PCA jittering as well. Furthermore, the selection of dataset was explained by the help of plots to prove the significance of extending the dataset. A new CNN model named LeafNet was introduced in [ 88 ] to classify the tea leaf diseases and achieved higher accuracy than Support Vector Machine (SVM) and Multi-Layer Perceptron (MLP). In [ 89 ], two DL models named modified MobileNet and reduced MobileNet were introduced, and their accuracy was near to the VGG model; the reduced MobileNet actually got 98.34% classification accuracy and had a fewer number of parameters as compared to VGG which saves time in training the model. A state-of-the-art DL model was proposed in [ 90 ] named PlantdiseaseNet which was remarkably suitable for the complex environment of an agricultural field. In [ 85 ], five types of apple plant diseases were classified and detected by the state-of-the-art CNN model named VGG-inception architecture. It outclassed the performance of many DL architectures like AlexNet, GoogLeNet, several versions of ResNet, and VGG. It also presented inter object/class detection and activation visualization; it was also mentioned for its clear vision of diseases in the plants.
A bar chart presented in Figure 12 indicates, from the most to the least frequently used, DL models for plant disease detection and classification. It can be clearly seen that the AlexNet model has been used in most of the researches. GoogLeNet, VGG-16, and ResNet-50 are the next most commonly used DL models. Similarly, there are some improved/cascaded versions (Improved Cifar-10, VGG-Inception, Cascaded AlexNet with GoogLeNet, reduced/modified MobileNet, modified LeNet, and modified GoogLeNet), which have been used for plant disease identification.

Deep learning models used in the particular number of research papers.
Summing up Section 2 , all the DL approaches along with the selected plant species and performance metrics are shown in Table 3 .
Comparison of several DL approaches in terms of various performance metrics.
| DL Architectures/Algorithms | Datasets | Selected Plant/s | Performance Metrics (and Their Results) | Refs |
|---|---|---|---|---|
| CNN | PlantVillage | Maize | CA (92.85%) | [ ] |
| AlexNet, GoogLeNet, ResNet | PlantVillage | Tomato | CA by ResNet which gave the best value (97.28%) | [ ] |
| LeNet | PlantVillage | Banana | CA (98.61%), F1 (98.64%) | [ ] |
| AlexNet, ALexNetOWTBn, GoogLeNet, Overfeat, VGG | PlantVillage and in-field images | Apple, blueberry, banana, cabbage, cassava, cantaloupe, celery, cherry, cucumber, corn, eggplant, gourd, grape, orange, onion | Success rate of VGG (99.53%) which is the best among all | [ ] |
| AlexNet, VGG16, VGG 19, SqueezeNet, GoogLeNet, Inceptionv3, InceptionResNetv2, ResNet50, Resnet101 | Real field dataset | Apricot, Walnut, Peach, Cherry | F1(97.14), Accuracy (97.86 ± 1.56) of ResNet | [ ] |
| Inceptionv3 | Experimental field dataset | Cassava | CA (93%) | [ ] |
| CNN | Images taken from the research center | Cucumber | CA (82.3%) | [ ] |
| Super-Resolution Convolutional Neural Network (SCRNN) | PlantVillage | Tomato | Accuracy (~90%) | [ ] |
| CaffeNet | Downloaded from the internet | Pear, cherry, peach, apple, grapevine | Precision (96.3%) | [ ] |
| AlexNet and GoogLeNet | PlantVillage | Apple, blueberry, bell pepper, cherry, corn, peach, grape, raspberry, potato, squash, soybean, strawberry, tomato | CA (99.35%) of GoogLeNet | [ ] |
| AlexNet, GoogLeNet, VGG- 16, ResNet-50,101, ResNetXt-101, Faster RCNN, SSD, R-FCN, ZFNet | Image taken in real fields | Tomato | Precision (85.98%) of ResNet-50 with Region based Fully Convolutional Network(R-FCN) | [ ] |
| CNN | Bisque platform of Cy Verse | Maize | Accuracy (96.7%) | [ ] |
| DCNN | Images were taken in real field | Rice | Accuracy (95.48%) | [ ] |
| AlexNet, GoogLeNet | PlantVillage | Tomato | Accuracy (0.9918 ± 0.169) of GoogLeNet | [ ] |
| VGG-FCN-VD16 and VGG-FCN-S | Wheat Disease Database 2017 | Wheat | Accuracy (97.95%) of VGG-FCN-VD16 | [ ] |
| VGG-A, CNN | Images were taken in real field | Radish | Accuracy (93.3%) | [ ] |
| AlexNet | Images were taken in real field | Soybean | CA (94.13%) | [ ] |
| AlexNet and SqueezeNet v1.1 | PlantVillage | Tomato | CA (95.65%) of AlexNet | [ ] |
| DCNN, Random forest, Support Vector Machine and AlexNet | PlantVillage dataset, Forestry Image dataset and agricultural field in China | Cucumber | CA (93.4%) of DCNN | [ ] |
| Teacher/student architecture | PlantVillage | Apple, bell pepper, blueberry, cherry, corn, orange, grape, potato, raspberry, peach, soybean, strawberry, tomato, squash | Training accuracy and loss (~99%,~0–0.5%), validation accuracy and loss (~95%, ~10%) | [ ] |
| Improved GoogLeNet, Cifar-10 | PlantVillage and various websites | Maize | Top-1 accuracy (98.9%) of improved GoogLeNet | [ ] |
| MobileNet, Modified MobileNet, Reduced MobileNet | PlantVillage dataset | 24 types of plant | CA (98.34%) of reduced MobileNet | [ ] |
| VGG-16, ResNet-50,101,152, Inception-V4 and DenseNets-121 | PlantVillage | Apple, bell pepper, blueberry, cherry, corn, orange, grape, potato, raspberry, peach, soybean, strawberry, tomato, squash | Testing accuracy (99.75%) of DenseNets | [ ] |
| User defined CNN, SVM, AlexNet, GoogLeNet, ResNet-20 and VGG-16 | Images were taken in real field | Apple | CA (97.62%) of proposed CNN | [ ] |
| AlexNet and VGG-16 | PlantVillage | Tomato | CA (AlexNet) | [ ] |
| LeafNet, SVM, MLP | Images were taken in real field | Tea leaf | CA (90.16%) of LeafNet | [ ] |
| 2D-CNN-BidGRU | Real wheat field | wheat | F1 (0.75) and accuracy (0.743) | [ ] |
| OR-AC-GAN | Real environment | Tomato | Accuracy (96.25%) | [ ] |
| 3D CNN | Real environment | Soybean | CA (95.73%), F1-score (0.87) | [ ] |
| DCNN | Real environment | Wheat | Accuracy (85%) | [ ] |
| ResNet-50 | Real environment | Wheat | Balanced Accuracy (87%) | [ ] |
| GPDCNN | Real environment | Cucumber | CA (94.65%) | [ ] |
| VGG-16, AlexNet | PlantVillage, CASC-IFW | Apple, banana | CA (98.6%) | [ ] |
| LeNet | Real environment | Grapes | CA (95.8%) | [ ] |
| PlantDiseaseNet | Real environment | Apple, bell-pepper, cherry, grapes, onion, peach, potato, plum, strawberry, sugar-beets, tomato, wheat | CA (93.67%) | [ ] |
| LeNet | PlantVillage | Soybean | CA (99.32%) | [ ] |
| VGG-Inception | Real environment | Apple | Mean average accuracy (78.8%) | [ ] |
| Resnet-50, Inception-V2, MobileNet-V1 | Real environment | Banana | Mean average accuracy (99%) of ResNet-50 | [ ] |
| Modified LeNet | PlantVillage | Olives | True positive rate (98.6 ± 1.47%) | [ ] |
3. Hyper-Spectral Imaging with DL Models
For early detection of plant diseases, several imaging techniques like multispectral imaging [ 91 ], thermal imaging, fluorescence and hyperspectral imaging are used [ 92 ]. Among them, hyperspectral imaging (HSI) is the focus of recent research. For example, [ 93 ] used hyperspectral imaging (HSI) to detect tomato plant diseases by identifying the region of interest, and a feature ranking-KNN (FR-KNN) model produced a satisfactory result for the detection of diseased and healthy plants. In the recent approach, HSI was used for the detection of an apple disease. Moreover, the redundancy issue was resolved by an unsupervised feature selection procedure known as Orthogonal Subspace Projection [ 94 ]. In [ 95 ], leaf diseases on peanuts were detected by HSI by identifying sensitive bands and hyperspectral vegetation index. The tomato disease detection was done by SVM classifiers based on HSI, and their performance was evaluated by F1-score, accuracy, specificity, and sensitivity [ 96 ].
Recently, HSI has been used with machine learning (ML) for the detection of plant diseases. For example, [ 97 ] described ML techniques for hyperspectral imaging for many agricultural applications. Moreover, ML with HSI have been used for three ML models, implemented by using hyperspectral measurement technique for the detection of leaf rust disease [ 98 ]. For wheat disease detection, [ 99 ] used Random Forest (RF) classifier with multispectral imaging technique and achieved accuracy of 89.3%. Plants’ diseases were also detected by SVM based on hyperspectral data and achieved accuracy of more than 86% [ 100 ]. There are some other ML approaches based on HSI [ 101 ], but this review is focused on DL approaches based on HSI, presented below.
The DL has been used to classify the hyperspectral images for many applications. For medical purposes, this technology is very useful as it is used for the classification of head/neck cancer in [ 102 ]. In [ 103 ], a DL approach based on HSI was proposed through contextual information as it provides spectral and spatial features. A new 3D-CNN architecture allowed for a fast, accurate, and efficient approach to classify the hyperspectral images in [ 104 ]. This architecture not only used the spectral information (as used in previous CNN techniques [ 105 ]) but also ensured that the spatial information was also taken into account. In [ 106 ], the feature extraction procedure was used with CNN for hyperspectral image classification and used dropout and L2 regularization methods in order to prevent overfitting. Just like CNN models used for hyperspectral imaging classification, RNN models are also used with HSI as described in [ 107 , 108 ]. In the domain of plant disease detection, some researches utilized Hyperspectral Imaging (HSI) along with DL models to observe clearer vision for symptoms of plant diseases. A hybrid method to classify the hyperspectral images was proposed in [ 109 ] consisting of DCNN, LR, and PCA and got better results compared to the previous methods for classification tasks. In [ 110 ], a detailed review of DL with HSI technique was provided. In order to avoid the overfitting and improve accuracy, a detailed comparison provided between several DL models like 1D/2D-CNN (2D-CNN better result), LSTM/GRU (both faced overfitting), 2D-CNN-LSTM/GRU (still overfitting) was observed. Therefore, a new hybrid approach from Convolutional and Bidirectional Gated Recurrent Network named 2D-CNN-BidLSTM/GRU was proposed for the hyperspectral images, which resolved the problem of overfitting and achieved 0.75 F1-score and 0.73 accuracy for wheat diseases detection [ 111 ]. According to [ 112 ], a hyperspectral proximal-sensing procedure based on the newest DL technique named Generative Adversarial Nets (GAN) was proposed in order to detect tomato plant disease before its clear symptoms appeared (as shown in Figure 13 ). In [ 84 ], a 3D-CNN approach was proposed for hyperspectral images to identify the Charcoal rot disease in soybeans and the CNN model was evaluated by accuracy (95.76%) and F1-score (0.87). The saliency map visualization was used, and the most delicate wavelength resulted as 733 nm, which approximately lies in the region of the wavelength of NIR. For the detection of potato virus, [ 113 ] described it by DL on the hyperspectral images and achieved acceptable values of precision (0.78) and recall (0.88). In [ 114 ], a DL model named multiple Inception-Resnet model was developed by using both spatial and spectral data on hyperspectral UAV images to detect the yellow rust in wheat (as shown in Figure 14 ). This model achieved an 85% accuracy, which is quite a lot higher than the RF-classifier (77%).

Sample images of OR-AC-GAN (a hyperspectral imaging model) [ 112 ].

Hyperspectral images by UAV: ( a ) RGB color plots, ( b ) Random-Forest classifier, and ( c ) proposed multiple Inception-ResNet model [ 114 ].
From this section, we can conclude that, although there are some DL models/architectures developed for hyperspectral image classification in the application of plant disease detection, this is still a fertile area of research and should lead to improvements for better detection of plants’ diseases [ 115 ] in different situations, like various conditions of illumination, considering real background, etc.
In Figure 13 , the resultant images are taken from the proposed method described in [ 112 ]. The green-colored portion indicates the healthy part of the plant; the red portion denotes the infected portion. Note that ( a ) and ( b ) are the healthy plant images as there is no red color indication, whereas ( c ) has infected disease which can be seen in its corresponding figure ( d ).
A comparison of proposed DCNN with RF classifier and RGB colored hyperspectral images are shown in Figure 14 . The red color label indicates the portion infected by rust. It should be observed that the rust plots were identified in an almost similar manner (see (b) and (c) of first row), but in the healthy plot, there was a large portion covered by the red label in (b) as compared to (c), which shows a wrong classification by RF model [ 114 ].
4. Conclusions and Future Directions
This review explained DL approaches for the detection of plant diseases. Moreover, many visualization techniques/mappings were summarized to recognize the symptoms of diseases. Although much significant progress was observed during the last three to four years, there are still some research gaps which are described below:
- In most of the researches (as described in the previous sections), the PlantVillage dataset was used to evaluate the accuracy and performance of the respective DL models/architectures. Although this dataset has a lot of images of several plant species with their diseases, it has a simple/plain background. However, for a practical scenario, the real environment should be considered.
- Hyperspectral/multispectral imaging is an emerging technology and has been used in many areas of research (as described in Section 3 ). Therefore, it should be used with the efficient DL architectures to detect the plants’ diseases even before their symptoms are clearly apparent.
- A more efficient way of visualizing the spots of disease in plants should be introduced as it will save costs by avoiding the unnecessary application of fungicide/pesticide/herbicide.
- The severity of plant diseases changes with the passage of time, therefore, DL models should be improved/modified to enable them to detect and classify diseases during their complete cycle of occurrence.
- DL model/architecture should be efficient for many illumination conditions, so the datasets should not only indicate the real environment but also contain images taken in different field scenarios.
- A comprehensive study is required to understand the factors affecting the detection of plant diseases, like the classes and size of datasets, learning rate, illumination, and the like.
Abbreviations
The abbreviations used in this manuscript are given as under:
| ML | Machine Learning |
| DL | Deep Learning |
| CNN | Convolutional Neural network |
| DCNN | Deep Convolutional Neural Network |
| ILSVRC | ImageNet Large Scale Visual Recognition Challenge |
| RF | Random Forest |
| CA | Classification Accuracy |
| LSTM | Long Short-Term Memory |
| IoU | Intersection of Union |
| NiN | Network in Network |
| RCN | Region based Convolutional Neural Network |
| FCN | Fully Convolutional Neural Network |
| YOLO | You Only Look Once |
| SSD | Single Shot Detector |
| PSPNet | Pyramid Scene Parsing Network |
| IRRCNN | Inception Recurrent Residual Convolutional Neural Network |
| IRCNN | Inception Recurrent Convolutional Neural Network |
| DCRN | Densely Connected Recurrent Convolutional Network |
| INAR-SSD | Single Shot Detector with Inception module and Rainbow concatenation |
| R2U-Net | Recurrent Residual Convolutional Neural Network based on U-Net model |
| SVM | Support Vector Machines |
| ELM | Extreme Learning Machine |
| KNN | K-Nearest Neighbor |
| SRCNN | Super-Resolution Convolutional Neural Network |
| R-FCN | Region-based Fully Convolutional Networks |
| ROC | Receiver Operating Characteristic |
| PCA | Principal Component Analysis |
| MLP | Multi-Layer Perceptron |
| LRP | Layer-wise Relevance Propagation |
| HSI | Hyperspectral Imaging |
| FRKNN | Feature Ranking K-Nearest Neighbor |
| RNN | Recurrent Neural Network |
| ToF | Time-of-Flight |
| LR | Logistic Regression |
| GRU | Gated Recurrent Unit |
| AN | Generative Adversarial Nets |
| GPDCNN | Global Pooling Dilated Convolutional Neural Network |
| 2D-CNN-BidGRU | 2D-Convolutional-Bidirectional Gated Recurrent Unit Neural Network |
| OR-AC-GAN | Outlier Removal-Auxiliary Classifier-Generative Adversarial Nets |
Author Contributions
Conceptualization, M.H.S. and K.M.A.; methodology, M.H.S. and K.M.A.; writing—original draft preparation, M.H.S. and K.M.A.; writing—review and editing, M.H.S., J.P., and K.M.A; visualization, M.H.S., J.P., and K.M.A; supervision, J.P., and K.M.A.; project administration, J.P., and K.M.A.
This research was funded by the Ministry of Business, Innovation and Employment (MBIE), New Zealand, Science for Technological Innovation (SfTI) National Science Challenge.
Conflicts of Interest
The authors declare no conflict of interest.
IEEE Account
- Change Username/Password
- Update Address
Purchase Details
- Payment Options
- Order History
- View Purchased Documents
Profile Information
- Communications Preferences
- Profession and Education
- Technical Interests
- US & Canada: +1 800 678 4333
- Worldwide: +1 732 981 0060
- Contact & Support
- About IEEE Xplore
- Accessibility
- Terms of Use
- Nondiscrimination Policy
- Privacy & Opting Out of Cookies
A not-for-profit organization, IEEE is the world's largest technical professional organization dedicated to advancing technology for the benefit of humanity. © Copyright 2024 IEEE - All rights reserved. Use of this web site signifies your agreement to the terms and conditions.
- Open access
- Published: 24 February 2021
Plant diseases and pests detection based on deep learning: a review
- Jun Liu ORCID: orcid.org/0000-0001-8769-5981 1 &
- Xuewei Wang 1
Plant Methods volume 17 , Article number: 22 ( 2021 ) Cite this article
129k Accesses
364 Citations
17 Altmetric
Metrics details
Plant diseases and pests are important factors determining the yield and quality of plants. Plant diseases and pests identification can be carried out by means of digital image processing. In recent years, deep learning has made breakthroughs in the field of digital image processing, far superior to traditional methods. How to use deep learning technology to study plant diseases and pests identification has become a research issue of great concern to researchers. This review provides a definition of plant diseases and pests detection problem, puts forward a comparison with traditional plant diseases and pests detection methods. According to the difference of network structure, this study outlines the research on plant diseases and pests detection based on deep learning in recent years from three aspects of classification network, detection network and segmentation network, and the advantages and disadvantages of each method are summarized. Common datasets are introduced, and the performance of existing studies is compared. On this basis, this study discusses possible challenges in practical applications of plant diseases and pests detection based on deep learning. In addition, possible solutions and research ideas are proposed for the challenges, and several suggestions are given. Finally, this study gives the analysis and prospect of the future trend of plant diseases and pests detection based on deep learning.
Plant diseases and pests detection is a very important research content in the field of machine vision. It is a technology that uses machine vision equipment to acquire images to judge whether there are diseases and pests in the collected plant images [ 1 ]. At present, machine vision-based plant diseases and pests detection equipment has been initially applied in agriculture and has replaced the traditional naked eye identification to some extent.
For traditional machine vision-based plant diseases and pests detection method, conventional image processing algorithms or manual design of features plus classifiers are often used [ 2 ]. This kind of method usually makes use of the different properties of plant diseases and pests to design the imaging scheme and chooses appropriate light source and shooting angle, which is helpful to obtain images with uniform illumination. Although carefully constructed imaging schemes can greatly reduce the difficulty of classical algorithm design, but also increase the application cost. At the same time, under natural environment, it is often unrealistic to expect the classical algorithms designed to completely eliminate the impact of scene changes on the recognition results [ 3 ]. In real complex natural environment, plant diseases and pests detection is faced with many challenges, such as small difference between the lesion area and the background, low contrast, large variations in the scale of the lesion area and various types, and a lot of noise in the lesion image. Also, there are a lot of disturbances when collecting plant diseases and pests images under natural light conditions. At this time, the traditional classical methods often appear helpless, and it is difficult to achieve better detection results.
In recent years, with the successful application of deep learning model represented by convolutional neural network (CNN) in many fields of computer vision (CV, computer-vision), for example, traffic detection [ 4 ], medical Image Recognition [ 5 ], Scenario text detection [ 6 ], expression recognition [ 7 ], face Recognition [ 8 ], etc. Several plant diseases and pests detection methods based on deep learning are applied in real agricultural practice, and some domestic and foreign companies have developed a variety of deep learning-based plant diseases and pests detection Wechat applet and photo recognition APP software. Therefore, plant diseases and pests detection method based on deep learning not only has important academic research value, but also has a very broad market application prospect.
In view of the lack of comprehensive and detailed discussion on plant diseases and pests detection methods based on deep learning, this study summarizes and combs the relevant literatures from 2014 to 2020, aiming to help researchers quickly and systematically understand the relevant methods and technologies in this field. The content of this study is arranged as follows: “ Definition of plant diseases and pests detection problem ” section gives the definition of plant diseases and pests detection problem; “ Image recognition technology based on deep learning ” section focuses on the detailed introduction of image recognition technology based on deep learning; “ Plant diseases and pests detection methods based on deep learning ” section analyses the three kinds of plant diseases and pests detection methods based on deep learning according to network structure, including classification, detection and segmentation network; “ Dataset and performance comparison ” section introduces some datasets of plant diseases and pests detection and compares the performance of the existing studies; “ Challenges ” section puts forward the challenges of plant diseases and pests detection based on deep learning; “ Conclusions and future directions ” section prospects the possible research focus and development direction in the future.
Definition of plant diseases and pests detection problem
Definition of plant diseases and pests
Plant diseases and pests is one kind of natural disasters that affect the normal growth of plants and even cause plant death during the whole growth process of plants from seed development to seedling and to seedling growth. In machine vision tasks, plant diseases and pests tend to be the concepts of human experience rather than a purely mathematical definition.
Definition of plant diseases and pests detection
Compared with the definite classification, detection and segmentation tasks in computer vision [ 9 ], the requirements of plant diseases and pests detection is very general. In fact, its requirements can be divided into three different levels: what, where and how [ 10 ]. In the first stage, “what” corresponds to the classification task in computer vision. As shown in Fig. 1 , the label of the category to which it belongs is given. The task in this stage can be called classification and only gives the category information of the image. In the second stage, “where” corresponds to the location task in computer vision, and the positioning of this stage is the rigorous sense of detection. This stage not only acquires what types of diseases and pests exist in the image, but also gives their specific locations. As shown in Fig. 1 , the plaque area of gray mold is marked with a rectangular box. In the third stage, “how” corresponds to the segmentation task in computer vision. As shown in Fig. 1 , the lesions of gray mold are separated from the background pixel by pixel, and a series of information such as the length, area, location of the lesions of gray mold can be further obtained, which can assist the higher-level severity level evaluation of plant diseases and pests. Classification describes the image globally through feature expression, and then determines whether there is a certain kind of object in the image by means of classification operation; while object detection focuses on local description, that is, answering what object exists in what position in an image, so in addition to feature expression, object structure is the most obvious feature that object detection differs from object classification. That is, feature expression is the main research line of object classification, while structure learning is the research focus of object detection. Although the function requirements and objectives of the three stages of plant diseases and pests detection are different, yet in fact, the three stages are mutually inclusive and can be converted. For example, the “where” in the second stage contains the process of “what” in the first stage, and the “how” in the third stage can finish the task of “where” in the second stage. Also, the “what” in the first stage can achieve the goal of the second and the third stages through some methods. Therefore, the problem in this study is collectively referred to as plant diseases and pests detection as conventions in the following text, and the terminology differentiates only when different network structures and functions are adopted.
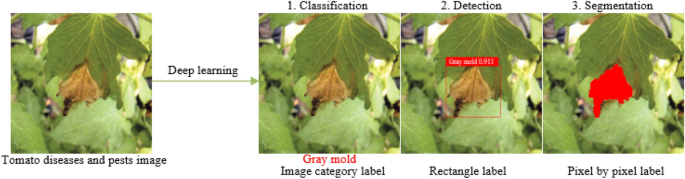
Comparison with traditional plant diseases and pests detection methods
To better illustrate the characteristics of plant diseases and pests detection methods based on deep learning, according to existing references [ 11 , 12 , 13 , 14 , 15 ], a comparison with traditional plant diseases and pests detection methods is given from four aspects including essence, method, required conditions and applicable scenarios. Detailed comparison results are shown in Table 1 .
Image recognition technology based on deep learning
Compared with other image recognition methods, the image recognition technology based on deep learning does not need to extract specific features, and only through iterative learning can find appropriate features, which can acquire global and contextual features of images, and has strong robustness and higher recognition accuracy.
Deep learning theory
The concept of Deep Learning (DL) originated from a paper published in Science by Hinton et al. [ 16 ] in 2006. The basic idea of deep learning is: using neural network for data analysis and feature learning, data features are extracted by multiple hidden layers, each hidden layer can be regarded as a perceptron, the perceptron is used to extract low-level features, and then combine low-level features to obtain abstract high-level features, which can significantly alleviate the problem of local minimum. Deep learning overcomes the disadvantage that traditional algorithms rely on artificially designed features and has attracted more and more researchers’ attention. It has now been successfully applied in computer vision, pattern recognition, speech recognition, natural language processing and recommendation systems [ 17 ].
Traditional image classification and recognition methods of manual design features can only extract the underlying features, and it is difficult to extract the deep and complex image feature information [ 18 ]. And deep learning method can solve this bottleneck. It can directly conduct unsupervised learning from the original image to obtain multi-level image feature information such as low-level features, intermediate features and high-level semantic features. Traditional plant diseases and pests detection algorithms mainly adopt the image recognition method of manual designed features, which is difficult and depends on experience and luck, and cannot automatically learn and extract features from the original image. On the contrary, deep learning can automatically learn features from large data without manual manipulation. The model is composed of multiple layers, which has good autonomous learning ability and feature expression ability, and can automatically extract image features for image classification and recognition. Therefore, deep learning can play a great role in the field of plant diseases and pests image recognition. At present, deep learning methods have developed many well-known deep neural network models, including deep belief network (DBN), deep Boltzmann machine (DBM), stack de-noising autoencoder (SDAE) and deep convolutional neural network (CNN) [ 19 ]. In the area of image recognition, the use of these deep neural network models to realize automate feature extraction from high-dimensional feature space offers significant advantages over traditional manual design feature extraction methods. In addition, as the number of training samples grows and the computational power increases, the characterization power of deep neural networks is being further improved. Nowadays, the boom of deep learning is sweeping both industry and academia, and the performance of deep neural network models are all significantly ahead of traditional models. In recent years, the most popular deep learning framework is deep convolutional neural network.
- Convolutional neural network
Convolutional Neural Networks, abbreviated as CNN, has a complex network structure and can perform convolution operations. As shown in Fig. 2 , the convolutional neural network model is composed of input layer, convolution layer, pooling layer, full connection layer and output layer. In one model, the convolution layer and the pooling layer alternate several times, and when the neurons of the convolution layer are connected to the neurons of the pooling layer, no full connection is required. CNN is a popular model in the field of deep learning. The reason lies in the huge model capacity and complex information brought about by the basic structural characteristics of CNN, which enables CNN to play an advantage in image recognition. At the same time, the successes of CNN in computer vision tasks have boosted the growing popularity of deep learning.
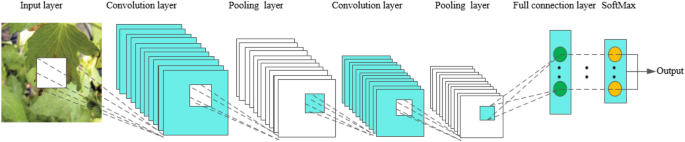
The basic structure of CNN
In the convolution layer, a convolution core is defined first. The convolution core can be considered as a local receptive field, and the local receptive field is the greatest advantage of the convolution neural network. When processing data information, the convolution core slides on the feature map to extract part of the feature information. After the feature extraction of the convolution layer, the neurons are input into the pooling layer to extract the feature again. At present, the commonly used methods of pooling include calculating the mean, maximum and random values of all values in the local receptive field [ 20 , 21 ]. After the data entering several convolution layers and pooling layers, they enter the full-connection layer, and the neurons in the full-connection layer are fully connected with the neurons in the upper layer. Finally, the data in the full-connection layer can be classified by the softmax method, and then the values are transmitted to the output layer for output results.
Open source tools for deep learning
The commonly used third-party open source tools for deep learning are Tensorflow [ 22 ], Torch/PyTorch [ 23 ], Caffe [ 24 ], Theano [ 25 ]. The different characteristics of each open source tool are shown in Table 2 .
The four commonly used deep learning third-party open source tools all support cross-platform operation, and the platforms that can be run include Linux, Windows, iOS, Android, etc. Torch/PyTorch and Tensorflow have good scalability and support a large number of third-party libraries and deep network structures, and have the fastest training speed when training large CNN networks on GPU.
Plant diseases and pests detection methods based on deep learning
This section gives a summary overview of plant diseases and pests detection methods based on deep learning. Since the goal achieved is completely consistent with the computer vision task, plant diseases and pests detection methods based on deep learning can be seen as an application of relevant classical networks in the field of agriculture. As shown in Fig. 3 , the network can be further subdivided into classification network, detection network and segmentation network according to the different network structures. As can be seen from Fig. 3 , this paper is subdivided into several different sub-methods according to the processing characteristics of each type of methods.
Framework of plant diseases and pests detection methods based on deep learning
Classification network
In real natural environment, the great differences in shape, size, texture, color, background, layout and imaging illumination of plant diseases and pests make the recognition a difficult task. Due to the strong feature extraction capability of CNN, the adoption of CNN-based classification network has become the most commonly used pattern in plant diseases and pests classification. Generally, the feature extraction part of CNN classification network consists of cascaded convolution layer + pooling layer, followed by full connection layer (or average pooling layer) + softmax structure for classification. Existing plant diseases and pests classification network mostly use the muture network structures in computer vision, including AlexNet [ 26 ], GoogleLeNet [ 27 ], VGGNet [ 28 ], ResNet [ 29 ], Inception V4 [ 30 ], DenseNets [ 31 ], MobileNet [ 32 ] and SqueezeNet [ 33 ]. There are also some studies which have designed network structures based on practical problems [ 34 , 35 , 36 , 37 ]. By inputting a test image into the classification network, the network analyses the input image and returns a label that classifies the image. According to the difference of tasks achieved by the classification network method, it can be subdivided into three subcategories: using the network as a feature extractor, using the network for classification directly and using the network for lesions location.
Using network as feature extractor
In the early studies on plant diseases and pests classification methods based on deep learning, many researchers took advantage of the powerful feature extraction capability of CNN, and the methods were combined with traditional classifiers [ 38 ]. First, the images are input into a pretrained CNN network to obtain image characterization features, and the acquired features are then input into a conventional machine learning classifier (e.g., SVM) for classification. Yalcin et al. [ 39 ] proposed a convolutional neural network architecture to extract the features of images while performing experiments using SVM classifiers with different kernels and feature descriptors such as LBP and GIST, the experimental results confirmed the effectiveness of the approach. Fuentes et al. [ 40 ] put forward the idea of CNN based meta architecture with different feature extractors, and the input images included healthy and infected plants, which were identified as their respective classes after going through the meta architecture. Hasan et al. [ 41 ] identified and classified nine different types of rice diseases by using the features extracted from DCNN model and input into SVM, and the accuracy achieved 97.5%.
Using network for classification directly
Directly using classification network to classify lesions is the earliest common means of CNN applied in plant diseases and pests detection. According to the characteristics of existing research work, it can be further subdivided into original image classification, classification after locating Region of Interest (ROI) and multi-category classification.
Original image classification. That is, directly put the collected complete plant diseases and pests image into the network for learning and training. Thenmozhi et al. [ 42 ] proposed an effective deep CNN model, and transfer learning is used to fine-tune the pre-training model. Insect species were classified on three public insect datasets with accuracy of 96.75%, 97.47% and 95.97%, respectively. Fang et al. [ 43 ] used ResNet50 in plant diseases and pests detection. The focus loss function was used instead of the standard cross-entropy loss function, and the Adam optimization method was used to identify the leaf disease grade, and the accuracy achieved 95.61%.
Classification after locating ROI. For the whole image acquired, we should focus on whether there is a lesion in a fixed area, so we often obtain the region of interest (ROI) in advance, and then input the ROI into the network to judge the category of diseases and pests. Nagasubramanian et al. [ 44 ] used a new three-dimensional deep convolution neural network (DCNN) and salience map visualization method to identify healthy and infected samples of soybean stem rot, and the classification accuracy achieved 95.73%.
Multi-category classification. When the number of plant diseases and pests class to be classified exceed 2 class, the conventional plant diseases and pests classification network is the same as the original image classification method, that is, the output nodes of the network are the number of plant diseases and pests class + 1 (including normal class). However, multi-category classification methods often use a basic network to classify lesions and normal samples, and then share feature extraction parts on the same network to modify or increase the classification branches of lesion categories. This approach is equivalent to preparing a pre-training weight parameter for subsequent multi-objective plant diseases and pests classification network, which is obtained by binary training between normal samples and plant diseases and pests samples. Picon et al. [ 45 ] proposed a CNN architecture to identify 17 diseases in 5 crops, which seamlessly integrates context metadata, allowing training of a single multi-crop model. The model can achieve the following goals: (a) obtains richer and more robust shared visual features than the corresponding single crop; (b) is not affected by different diseases in which different crops have similar symptoms; (c) seamlessly integrates context to perform crop conditional disease classification. Experiments show that the proposed model alleviates the problem of data imbalance, and the average balanced accuracy is 0.98, which is superior to other methods and eliminates 71% of classifier errors.
Using network for lesions location
Generally, the classification network can only complete the classification of image label level. In fact, it can also achieve the location of lesions and the pixel-by-pixel classification by combining different techniques and methods. According to the different means used, it can be further divided into three forms: sliding window, heatmap and multi-task learning network.
Sliding window. This is the simplest and intuitive method to achieve the location of lesion coarsely. The image in the sliding window is input into the classification network for plant diseases and pests detection by redundant sliding on the original image through a smaller size window. Finally, all sliding windows are connected to obtain the results of the location of lesion. Chen et al. [ 46 ] used CNN classification network based on sliding window to build a framework for characteristics automatic learning, feature fusion, recognition and location regression calculation of plant diseases and pests species, and the recognition rate of 38 common symptoms in the field was 50–90%.
Heatmap. This is an image that reflects the importance of each region in the image, the darker the color represents the more important. In the field of plant diseases and pests detection, the darker the color in the heatmap represents the greater the probability that it is the lesion. In 2017, Dechant et al. [ 47 ] trained CNN to make heatmap to show the probability of infection in each region in maize disease images, and these heatmaps were used to classify the complete images, dividing each image into containing or not containing infected leaves. At runtime, it takes about 2 min to generate a heatmap for an image (1.6 GB of memory) and less than one second to classify a set of three heatmaps (800 MB of memory). Experiments show that the accuracy is 96.7% on the test dataset. In 2019, Wiesner-Hanks et al. [ 48 ] used heatmap method to obtain accurate contour areas of maize diseases, the model can accurately depict lesions as low as millimeter scale from the images collected by UAVs, with an accuracy rate of 99.79%, which is the best scale of aerial plant disease detection achieved so far.
Multi-task learning network. If the pure classified network does not add any other skills, it could only realize the image level classification. Therefore, to accurately locate the location of plant diseases and pests, the designed network should often add an extra branch, and the two branches would share the results of the feature extracting. In this way, the network generally had the classification and segmentation output of the plant diseases and pests, forming a multi-task learning network. It takes into account the characteristics of both network. For segmentation network branches, each pixel in the image can be used as a training sample to train the network. Therefore, the multi-task learning network not only uses the segmentation branches to output the specific segmentation results of the lesions, but also greatly reduces the requirements of the classification network for samples. Ren et al. [ 49 ] constructed a Deconvolution-Guided VGNet (DGVGNet) model to identify plant leaf diseases which were easily disturbed by shadows, occlusions and light intensity. The deconvolution was used to guide the CNN classifier to focus on the real lesion sites. The test results show that the accuracy of disease class identification is 99.19%, the pixel accuracy of lesion segmentation is 94.66%, and the model has good robustness in occlusion, low light and other environments.
To sum up, the method based on classification network is widely used in practice, and many scholars have carried out application research on the classification of plant diseases and pests [ 50 , 51 , 52 , 53 ]. At the same time, different sub-methods have their own advantages and disadvantages, as shown in Table 3 .
Detection network
Object positioning is one of the most basic tasks in the field of computer vision. It is also the closest task to plant diseases and pests detections in the traditional sense. Its purpose is to obtain accurate location and category information of the object. At present, object detection methods based on deep learning emerge endlessly. Generally speaking, plant diseases and pests detection network based on deep learning can be divided into: two stage network represented by Faster R-CNN [ 54 ]; one stage network represented by SSD [ 55 ] and YOLO [ 56 , 57 , 58 ]. The main difference between the two networks is that the two-stage network needs to first generate a candidate box (proposal) that may contain the lesions, and then further execute the object detection process. In contrast, the one-stage network directly uses the features extracted in the network to predict the location and class of the lesions.
Plant diseases and pests detection based on two stages network
The basic process of two-stage detection network (Faster R-CNN) is to obtain the feature map of the input image through the backbone network first, then calculate the anchor box confidence using RPN and get the proposal. Then, input the feature map of the proposal area after ROIpooling to the network, fine-tune the initial detection results, and finally get the location and classification results of the lesions. Therefore, according to the characteristics of plant diseases and pests detection, common methods often improve on the backbone structure or its feature map, anchor ratio, ROIpooling and loss function. In 2017, Fuentes et al. [ 59 ] first used Faster R-CNN to locate tomato diseases and pests directly, combined with deep feature extractors such as VGG-Net and ResNet, the mAP value reached 85.98% in a dataset containing 5000 tomato diseases and pests of 9 categories. In 2019, Ozguven et al. [ 60 ] proposed a Faster R-CNN structure for automatic detection of beet leaf spot disease by changing the parameters of CNN model. 155 images were trained and tested. The results show that the overall correct classification rate of this method is 95.48%. Zhou et al. [ 61 ] presented a fast rice disease detection method based on the fusion of FCM-KM and Faster R-CNN. The application results of 3010 images showed that: the detection accuracy and time of rice blast, bacterial blight, and sheath blight are 96.71%/0.65 s, 97.53%/0.82 s and 98.26%/0.53 s respectively. Xie et al. [ 62 ] proposed a Faster DR-IACNN model based on the self-built grape leaf disease dataset (GLDD) and Faster R-CNN detection algorithm, the Inception-v1 module, Inception-ResNet-v2 module and SE are introduced. The proposed model achieved higher feature extraction ability, the mAP accuracy was 81.1% and the detection speed was 15.01FPS. The two-stage detection network has been devoted to improving the detection speed to improve the real-time and practicability of the detection system, but compared with the single-stage detection network, it is still not concise enough, and the inference speed is still not fast enough.
Plant diseases and pests detection based on one stage network
The one-stage object detection algorithm has eliminated the region proposal stage, but directly adds the detection head to the backbone network for classification and regression, thus greatly improving the inference speed of the detection network. The single-stage detection network is divided into two types, SSD and YOLO, both of which use the whole image as the input of the network, and directly return the position of the bounding box and the category to which it belongs at the output layer.
Compared with the traditional convolutional neural network, the SSD selects VGG16 as the trunk of the network, and adds a feature pyramid network to obtain features from different layers and make predictions. Singh et al. [ 63 ] built the PlantDoc dataset for plant disease detection. Considering that the application should predict in mobile CPU in real time, an application based on MobileNets and SSD was established to simplify the detection of model parameters. Sun et al. [ 64 ] presented an instance detection method of multi-scale feature fusion based on convolutional neural network, which is improved on the basis of SSD to detect maize leaf blight under complex background. The proposed method combined data preprocessing, feature fusion, feature sharing, disease detection and other steps. The mAP of the new model is higher (from 71.80 to 91.83%) than that of the original SSD model. The FPS of the new model has also improved (from 24 to 28.4), reaching the standard of real-time detection.
YOLO considers the detection task as a regression problem, and uses global information to directly predict the bounding box and category of the object to achieve end-to-end detection of a single CNN network. YOLO can achieve global optimization and greatly improve the detection speed while satisfying higher accuracy. Prakruti et al. [ 65 ] presented a method to detect pests and diseases on images captured under uncontrolled conditions in tea gardens. YOLOv3 was used to detect pests and diseases. While ensuring real-time availability of the system, about 86% mAP was achieved with 50% IOU. Zhang et al. [ 66 ] combined the pooling of spatial pyramids with the improved YOLOv3, deconvolution is implemented by using the combination of up-sampling and convolution operation, which enables the algorithm to effectively detect small size crop pest samples in the image and reduces the problem of relatively low recognition accuracy due to the diversity of crop pest attitudes and scales. The average recognition accuracy can reach 88.07% by testing 20 class of pests collected in real scene.
In addition, there are many studies on using detection network to identify diseases and pests [ 47 , 67 , 68 , 69 , 70 , 71 , 72 , 73 ]. With the development of object detection network in computer vision, it is believed that more and more new detection models will be applied in plant diseases and pests detection in the future. In summary, in the field of plant diseases and pests detection which emphasizes detection accuracy at this stage, more models based on two-stage are used, and in the field of plant diseases and pests detection which pursue detection speed more models based on one-stage are used.
Can detection network replace classification network? The task of detection network is to solve the location problem of plant diseases and pests. The task of classification network is to judge the class of plant diseases and pests. Visually, the hidden information of detection network includes the category information, that is, the category information of plant diseases and pests that need to be located needs to be known beforehand, and the corresponding annotation information should be given in advance to judge the location of plant diseases and pests. From this point of view, the detection network seems to include the steps of the classification network, that is, the detection network can answer “what kind of plant diseases and pests are in what place”. But there is a misconception, in which “what kind of plant diseases and pests” is given a priori, that is, what is labelled during training is not necessarily the real result. In the case of strong model differentiation, that is, when the detection network can give accurate results, the detection network can answer “what kind of plant diseases and pests are in what place” to a certain extent. However, in the real world, in many cases, it cannot uniquely reflect the uniqueness of plant diseases and pests categories, only can answer “what kind of plant diseases and pests may be in what place”, then the involvement of the classification network is necessary. Thus, the detection network cannot replace the classification network.
Segmentation network
Segmentation network converts the plant diseases and pests detection task to semantic and even instance segmentation of lesions and normal areas. It not only finely divides the lesion area, but also obtains the location, category and corresponding geometric properties (including length, width, area, outline, center, etc.). It can be roughly divided into: Fully Convolutional Networks (FCN) [ 74 ] and Mask R-CNN [ 75 ].
Full convolution neural network (FCN) is the basis of image semantics segmentation. At present, almost all semantics segmentation models are based on FCN. FCN first extracts and codes the features of the input image using convolution, then gradually restores the feature image to the size of the input image by deconvolution or up sampling. Based on the differences in FCN network structure, the plant diseases and pests segmentation methods can be divided into conventional FCN, U-net [ 76 ] and SegNet [ 77 ].
Conventional FCN. Wang et al. [ 78 ] presented a new method of maize leaf disease segmentation based on full convolution neural network to solve the problem that traditional computer vision is susceptible to different illumination and complex background, and the segmentation accuracy reached 96.26. Wang et al. [ 79 ] proposed a plant diseases and pests segmentation method based on improved FCN. In this method, a convolution layer was used to extract multi-layer feature information from the input maize leaf lesion image, and the size and resolution of the input image were restored by deconvolution operation. Compared with the original FCN method, not only the integrity of the lesion was guaranteed, but also the segmentation of small lesion area was highlighted, and the accuracy rate reached 95.87%.
U-net. U-net is not only a classical FCN structure, but also a typical encoder-decoder structure. It is characterized by introducing a layer-hopping connection, fusing the feature map in the coding stage with that in the decoding stage, which is beneficial to the recovery of segmentation details. Lin et al. [ 80 ] used U-net based convolutional neural network to segment 50 cucumber powdery mildew leaves collected in natural environment. Compared with the original U-net, a batch normalization layer was added behind each convolution layer, making the neural network insensitive to weight initialization. The experiment shows that the convolutional neural network based on U-net can accurately segment powdery mildew on cucumber leaves at the pixel level with an average pixel accuracy of 96.08%, which is superior to the existing K-means, Random-forest and GBDT methods. The U-net method can segment the lesion area in a complex background, and still has good segmentation accuracy and segmentation speed with fewer samples.
SegNet. It is also a classical encoder–decoder structure. Its feature is that the up-sampling operation in the decoder takes advantage of the index of the largest pooling operation in the encoder. Kerkech et al. [ 81 ] presented an image segmentation method for unmanned aerial vehicles. Visible and infrared images (480 samples from each range) were segmented using SegNet to identify four categories: shadows, ground, healthy and symptomatic grape vines. The detection rates of the proposed method on grape vines and leaves were 92% and 87%, respectively.
Mask R-CNN is one of the most commonly used image instance segmentation methods at present. It can be considered as a multitask learning method based on detection and segmentation network. When multiple lesions of the same type have adhesion or overlap, instance segmentation can separate individual lesions and further count the number of lesions. However, semantic segmentation often treats multiple lesions of the same type as a whole. Stewart et al. [ 82 ] trained a Mask R-CNN model to segment maize northern leaf blight (NLB) lesions in an unmanned aerial vehicle image. The trained model can accurately detect and segment a single lesion. At the IOU threshold of 0.50, the IOU between the baseline true value and the predicted lesion was 0.73, and the average accuracy was 0.96. Also, some studies combine the Mask R-CNN framework with object detection networks for plant diseases and pests detection. Wang et al. [ 83 ] used two different models, Faster R-CNN and ask R-CNN, in which Faster R-CNN was used to identify the class of tomato diseases and Mask R-CNN was used to detect and segment the location and shape of the infected area. The results showed that the proposed model can quickly and accurately identify 11 class of tomato diseases, and divide the location and shape of infected areas. Mask R-CNN reached a high detection rate of 99.64% for all class of tomato diseases.
Compared with the classification and detection network methods, the segmentation method has advantages in obtaining the lesion information. However, like the detection network, it requires a lot of annotation data, and its annotation information is pixel by pixel, which often takes a lot of effort and cost.
Dataset and performance comparison
This section first gives a brief introduction to the plant diseases and pests related datasets and the evaluation index of deep learning model, then compares and analyses the related models of plant diseases and pests detection based on deep learning in recent years.
Datasets for plant diseases and pests detection
Plant diseases and pests detection datasets are the basis for research work. Compared with ImageNet, PASCAL-VOC2007/2012 and COCO in computer vision tasks, there is not a large and unified dataset for plant diseases and pests detection. The plant diseases and pests dataset can be acquired by self-collection, network collection and use of public datasets. Among them, self-collection of image dataset is often obtained by unmanned aerial remote sensing, ground camera photography, Internet of Things monitoring video or video recording, aerial photography of unmanned aerial vehicle with camera, hyperspectral imager, near-infrared spectrometer, and so on. Public datasets typically come from PlantVillage, an existing well-known public standard library. Relatively, self-collected datasets of plant diseases and pests in real natural environment are more practical. Although more and more researchers have opened up the images collected in the field, it is difficult to compare them uniformly based on different class of diseases under different detection objects and scenarios. This section provides links to a variety of plant diseases and pests detection datasets in conjunction with existing studies. As shown in Table 4 .
Evaluation indices
Evaluation indices can vary depending on the focus of the study. Common evaluation indices include \(Precision\) , \(Recall\) , mean Average Precision (mAP) and the harmonic Mean F1 score based on \(Precision\) and \(Recall\) .
\(Precision\) and \(Recall\) are defined as:
In Formula ( 1 ) and Formula ( 2 ), TP (True Positive) is true-positive, predicted to be 1 and actually 1, indicating the number of lesions correctly identified by the algorithm. FP (False Positive) is false-positive, predicted to be 1 and actually 0, indicating the number of lesions incorrectly identified by the algorithm. FN (False Negative) is false-negative, predicted to be 0 and actually 1, indicating the number of unrecognized lesions.
Detection accuracy is usually assessed using mAP. The average accuracy of each category in the dataset needs to be calculated first:
In the above-mentioned formula, \(N\left( {class} \right)\) represents the number of all categories, \(Precision\left( j \right)\) and \(Recall\left( j \right)\) represents the precision and recall of class j respectively.
Average accuracy for each category is defined as mAP:
The greater the value of \(mAP\) , the higher the recognition accuracy of the algorithm; conversely, the lower the accuracy of the algorithm.
F1 score is also introduced to measure the accuracy of the model. F1 score takes into account both the accuracy and recall of the model. The formula is
Frames per second (FPS) is used to evaluate the recognition speed. The more frames per second, the faster the algorithm recognition speed; conversely, the slower the algorithm recognition speed.
Performance comparison of existing algorithms
At present, the research on plant diseases and pests based on deep learning involves a wide range of crops, including all kinds of vegetables, fruits and food crops. The tasks completed include not only the basic tasks of classification, detection and segmentation, but also more complex tasks such as the judgment of infection degree.
At present, most of the current deep learning-based methods for plant diseases and pests detection are applied on specific datasets, many datasets are not publicly available, there is still no single publicly available and comprehensive dataset that will allow all algorithms to be uniformly compared. With the continuous development of deep learning, the application performance of some typical algorithms on different datasets has been gradually improved, and the mAP, F1 score and FPS of the algorithms have all been increased.
The breakthroughs achieved in the existing studies are amazing, but due to the fact that there is still a certain gap between the complexity of the infectious diseases and pests images in the existing studies and the real-time field diseases and pests detection based on mobile devices. Subsequent studies will need to find breakthroughs in larger, more complex, and more realistic datasets.
Small dataset size problem
At present, deep learning methods are widely used in various computer vision tasks, plant diseases and pests detection is generally regarded as specific application in the field of agriculture. There are too few agricultural plant diseases and pests samples available. Compared with open standard libraries, self-collected data sets are small in size and laborious in labeling data. Compared with more than 14 million sample data in ImageNet datasets, the most critical problem facing plant diseases and pests detection is the problem of small samples. In practice, some plant diseases have low incidence and high cost of disease image acquisition, resulting in only a few or dozen training data collected, which limits the application of deep learning methods in the field of plant diseases and pests identification. In fact, for the problem of small samples, there are currently three different solutions.
Data amplification, synthesis and generation
Data amplification is a key component of training deep learning models. An optimized data amplification strategy can effectively improve the plant diseases and pests detection effect. The most common method of plant diseases and pests image expansion is to acquire more samples using image processing operations such as mirroring, rotating, shifting, warping, filtering, contrast adjustment, and so on for the original plant diseases and pests samples. In addition, Generative Adversarial Networks (GANs) [ 93 ] and Variational automatic encoder (VAE) [ 94 ] can generate more diverse samples to enrich limited datasets.
Transfer learning and fine-tuning classical network model
Transfer learning (TL) transfers knowledge learned from generic large datasets to specialized areas with relatively small amounts of data. When transfer learning develops a model for newly collected unlabeled samples, it can start with a training model by a similar known dataset. After fine-tuning parameters or modifying components, it can be applied to localized plant disease and pest detection, which can reduce the cost of model training and enable the convolution neural network to adapt to small sample data. Oppenheim et al. [ 95 ] collected infected potato images of different sizes, hues and shapes under natural light and classified by fine-tuning the VGG network. The results showed that, the transfer learning and training of new networks were effective. Too et al. [ 96 ] evaluated various classical networks by fine-tuning and contrast. The experimental results showed that the accuracy of Dense-Nets improved with the number of iterations. Chen et al. [ 97 ] used transfer learning and fine-tuning to identify rice disease images under complex background conditions and achieved an average accuracy of 92.00%, which proves that the performance of transfer learning is better than training from scratch.
Reasonable network structure design
By designing a reasonable network structure, the sample requirements can be greatly reduced. Zhang et al. [ 98 ] constructed a three-channel convolution neural network model for plant leaf disease recognition by combining three color components. Each channel TCCNN component is composed of three color RGB leaf disease images. Liu et al. [ 99 ] presented an improved CNN method for identifying grape leaf diseases. The model used a depth-separable convolution instead of a standard convolution to alleviate overfitting and reduce the number of parameters. For the different size of grape leaf lesions, the initial structure was applied to the model to improve the ability of multi-scale feature extraction. Compared with the standard ResNet and GoogLeNet structures, this model has faster convergence speed and higher accuracy during training. The recognition accuracy of this algorithm was 97.22%.
Fine-grained identification of small-size lesions in early identification
Small-size lesions in early identification.
Accurate early detection of plant diseases is essential to maximize the yield [ 36 ]. In the actual early identification of plant diseases and pests, due to the small size of the lesion object itself, multiple down sampling processes in the deep feature extraction network tend to cause small-scale objects to be ignored. Moreover, due to the background noise problem on the collected images, large-scale complex background may lead to more false detection, especially on low-resolution images. In view of the shortage of existing algorithms, the improvement direction of small object detection algorithm is analyzed, and several strategies such as attention mechanism are proposed to improve the performance of small target detection.
The use of attention mechanism makes resources allocated more rationally. The essence of attention mechanism is to quickly find region of interest and ignore unimportant information. By learning the characteristics of plant diseases and pests images, features can be separated using weighted sum method with weighted coefficient, and the background noise in the image can be suppressed. Specifically, the attention mechanism module can get a salient image, and seclude the object from the background, and the Softmax function can be used to manipulate the feature image, and combine it with the original feature image to obtain new fusion features for noise reduction purposes. In future studies on early recognition of plant diseases and pests, attention mechanisms can be used to effectively select information and allocate more resources to region of interest to achieve more accurate detection. Karthik et al. [ 100 ] applied attention mechanism on the residual network and experiments were carried out using the plantVillage dataset, which achieved 98% overall accuracy.
Fine-grained identification
First, there is a large difference within the class, that is, the visual characteristics of plant diseases and pests belonging to the same class are quite different. The reason is that the aforementioned external factors such as uneven illumination, dense occlusion, blurred equipment dithering and other interferences, resulting in different image samples belonging to the same kind of diseases and pests differ greatly. Plant diseases and pests detection in complex scenarios is a very challenging task of fine-grained recognition [ 101 ]. The existence of growth variations of diseases and pests results in distinct differences in the characterization of the same diseases and pests at different stages, forming the “intra-class difference” fine-grained characteristics.
Secondly, there is fuzziness between classes, that is, objects of different classes have some similarity. There are many detailed classifications of biological subspecies and subclasses of different kinds of diseases and pests, and there are some similarities of biological morphology and life habits among the subclasses, which lead to the problem of fine-grained identification of “inter-class similarity”. Barbedo believed that similar symptoms could be produced, which even phytopathologists could not correctly distinguish [ 102 ].
Thirdly, background disturbance makes it impossible for plant diseases and pests to appear in a very clean background in the real world. Background can be very complex and interfere with objects of interest, which makes plant diseases and pests detection more difficult. Some literature often ignores this issue because images are captured under controlled conditions [ 103 ].
Relying on the existing deep learning methods can not effectively identify the fine-grained characteristics of diseases and pests that exist naturally in the application of the above actual agricultural scenarios, resulting in technical difficulties such as low identification accuracy and generalization robustness, which has long restricted the performance improvement of decision-making management of diseases and pests by the Intelligent Agricultural Internet of Things [ 104 ]. The existing research is only suitable for fine-grained identification of fewer class of diseases and pests, can not solve the problem of large-scale, large-category, accurate and efficient identification of diseases and pests, and is difficult to deploy directly to the mobile terminals of smart agriculture.
Detection performance under the influence of illumination and occlusion
Lighting problems.
Previous studies have collected images of plant diseases and pests mostly in indoor light boxes [ 105 ]. Although this method can effectively eliminate the influence of external light to simplify image processing, it is quite different from the images collected under real natural light. Because natural light changes very dynamically, and the range in which the camera can accept dynamic light sources is limited, it is easy to cause image color distortion when above or below this limit. In addition, due to the difference of view angle and distance during image collection, the apparent characteristics of plant diseases and pests change greatly, which brings great difficulties to the visual recognition algorithm.
Occlusion problem
At present, most researchers intentionally avoid the recognition of plant diseases and pests in complex environments. They only focus on a single background. They use the method of directly intercepting the area of interest to the collected images, but seldom consider the occlusion problem. As a result, the recognition accuracy under occlusion is low and the practicability is greatly reduced. Occlusion problems are common in real natural environments, including blade occlusion caused by changes in blade posture, branch occlusion, light occlusion caused by external lighting, and mixed occlusion caused by different types of occlusion. The difficulties of plant diseases and pests identification under occlusion are the lack of features and noise overlap caused by occlusion. Different occlusion conditions have different degrees of impact on the recognition algorithm, resulting in false detection or even missed detection. In recent years, with the maturity of deep learning algorithms under restricted conditions, some researchers have gradually challenged the identification of plant diseases and pests under occluded conditions [ 106 , 107 ], and significant progress has been made, which lays a good foundation for the application of plant diseases and pests identification in real-world scenarios. However, occlusion is random and complex. The training of the basic framework is difficult and the dependence on the performance of hardware devices still exists, we should strengthen the innovation and optimization of the basic framework, including the design of lightweight network architecture. The exploration of GAN and other aspects should be enhanced, while ensuring the accuracy of detection, the difficulty of model training should be reduced. GAN has prominent advantages in dealing with posture changes and chaotic background, but its design is not yet mature, and it is easy to crash in learning and cause model uncontrollable problems during training. We should strengthen the exploration of network performance to make it easier to quantify the quality of the model.
Detection speed problem
Compared with traditional methods, deep learning algorithms have better results, but their computational complexity is also higher. If the detection accuracy is guaranteed, the model needs to fully learn the characteristics of the image and increase the computational load, which will inevitably lead to slow detection speed and can not meet the needs of real-time. In order to ensure the detection speed, it is usually necessary to reduce the amount of calculation. However, this will cause insufficient training and result in false or missed detection. Therefore, it is important to design an efficient algorithm with both detection accuracy and detection speed.
Plant diseases and pests detection methods based on deep learning include three main links in agricultural applications: data labeling, model training and model inference. In real-time agricultural applications, more attention is paid to model inference. Currently, most plant diseases and pests detection methods focus on the accuracy of recognition. Little attention is paid to the efficiency of model inference. In reference [ 108 ], to improve the efficiency of the model calculation process to meet the actual agricultural needs, a deep separable convolution structure model for plant leaf disease detection was introduced. Several models were trained and tested. The classification accuracy of Reduced MobileNet was 98.34%, the parameters were 29 times less than VGG, and 6 times less than MobileNet. This shows an effective compromise between delay and accuracy, which is suitable for real-time crop diseases diagnosis on resource-constrained mobile devices.
Conclusions and future directions
Compared with traditional image processing methods, which deal with plant diseases and pests detection tasks in several steps and links, plant diseases and pests detection methods based on deep learning unify them into end-to-end feature extraction, which has a broad development prospects and great potential. Although plant diseases and pests detection technology is developing rapidly, it has been moving from academic research to agricultural application, there is still a certain distance from the mature application in the real natural environment, and there are still some problems to be solved.
Plant diseases and pests detection dataset
Deep learning technology has made some achievements in the identification of plant diseases and pests. Various image recognition algorithms have also been further developed and extended, which provides a theoretical basis for the identification of specific diseases and pests. However, the collection of image samples in previous studies mostly come from the characterization of disease spots, insect appearance characteristics or the characterization of insect pests and leaves. Most of the research results are limited to the laboratory environment and are applicable only to the plant diseases and pests images obtained at the time. The main reason for this is that the growth of plants is cyclical, continuous, seasonal and regional. Similarly, the characteristics of the same disease or pest at different growing stages of crops are different. Images of different plant species vary from region to region. As a result, most of the existing research results are not universal. Even with a high recognition rate in a single trial, the validity of the data obtained at other times cannot be guaranteed.
Most of the existing studies are based on the images generated in the visible range, but the electromagnetic wave outside the visible range also contains a lot of information, so the comprehensive information such as visible light, near infrared, multi-spectral should be fused to achieve the acquisition of plant diseases and pests dataset. Future research should focus on multi-information fusion method to obtain and identify plant diseases and pests information.
In addition, image databases of different kinds of plant diseases and pests in real natural environments are still in the blank stage. Future research should make full use of the data information acquisition platform such as portable field spore auto-capture instrument, unmanned aerial vehicle aerial photography system, agricultural internet of things monitoring equipment, which performs large-area and coverage identification of farmland and makes up for the lack of randomness of image samples in previous studies. Also, it can ensures the comprehensiveness and accuracy of dataset, and improves the generality of the algorithm.
Early recognition of plant diseases and pests
In the application of plant diseases and pests identification, the manifestation symptoms are not obvious, so early diagnosis is very difficult whether it is by visual observation or computer interpretation. However, the research significance and demand of early diagnosis are greater, which is more conducive to the prevention and control of plant diseases and pests and prevent their spread and development. The best image quality can be obtained when the sunlight is sufficient, and taking pictures in cloudy weather will increase the complexity of image preprocessing and reduce the recognition effect. In addition, in the early stage of plant diseases and pests occurrence, even high-resolution images are difficult to analyze. It is necessary to combine meteorological and plant protection data such as temperature and humidity to realize the recognition and prediction of diseases and pests. By consulting the existing research literatures, there are few reports on the early diagnosis of plant diseases and pests.
Network training and learning
When plant diseases and pests are visually identified manually, it is difficult to collect samples of all plant diseases and pests types, and many times only healthy data (positive samples) are available. However, most of the current plant diseases and pests detection methods based on deep learning are supervised learning based on a large number of diseases and pests samples, so manual collection of labelled datasets requires a lot of manpower, so unsupervised learning needs to be explored. Deep learning is a black box, which requires a large number of labelled training samples for end-to-end learning and has poor interpretability. Therefore, how to use the prior knowledge of brain-inspired computing and human-like visual cognitive model to guide the training and learning of the network is also a direction worthy of studying. At the same time, deep models need a large amount of memory and are extremely time-consuming during testing, which makes them unsuitable for deployment on mobile platforms with limited resources. It is important to study how to reduce complexity and obtain fast-executing models without losing accuracy. Finally, the selection of appropriate hyper-parameters has always been a major obstacle to the application of deep learning model to new tasks, such as learning rate, filter size, step size and number, these hyper-parameters have a strong internal dependence, any small adjustment may have a greater impact on the final training results.
Interdisciplinary research
Only by more closely integrating empirical data with theories such as agronomic plant protection, can we establish a field diagnosis model that is more in line with the rules of crop growth, and will further improve the effectiveness and accuracy of plant diseases and pests identification. In the future, it is necessary to go from image analysis at the surface level to identification of the occurrence mechanism of diseases and pests, and transition from simple experimental environment to practical application research that comprehensively considers crop growth law, environmental factors, etc.
In summary, with the development of artificial intelligence technology, the research focus of plant diseases and pests detection based on machine vision has shifted from classical image processing and machine learning methods to deep learning methods, which solved the difficult problems that could not be solved by traditional methods. There is still a long distance from the popularization of practical production and application, but this technology has great development potential and application value. To fully explore the potential of this technology, the joint efforts of experts from relevant disciplines are needed to effectively integrate the experience knowledge of agriculture and plant protection with deep learning algorithms and models, so as to make plant diseases and pests detection based on deep learning mature. Also, the research results should be integrated into agricultural machinery equipment to truly land the corresponding theoretical results.
Availability of data and materials
For relevant data and codes, please contact the corresponding author of this manuscript.
Lee SH, Chan CS, Mayo SJ, Remagnino P. How deep learning extracts and learns leaf features for plant classification. Pattern Recogn. 2017;71:1–13.
Article Google Scholar
Tsaftaris SA, Minervini M, Scharr H. Machine learning for plant phenotyping needs image processing. Trends Plant Sci. 2016;21(12):989–91.
Article CAS PubMed Google Scholar
Fuentes A, Yoon S, Park DS. Deep learning-based techniques for plant diseases recognition in real-field scenarios. In: Advanced concepts for intelligent vision systems. Cham: Springer; 2020.
Google Scholar
Yang D, Li S, Peng Z, Wang P, Wang J, Yang H. MF-CNN: traffic flow prediction using convolutional neural network and multi-features fusion. IEICE Trans Inf Syst. 2019;102(8):1526–36.
Sundararajan SK, Sankaragomathi B, Priya DS. Deep belief cnn feature representation based content based image retrieval for medical images. J Med Syst. 2019;43(6):1–9.
Melnyk P, You Z, Li K. A high-performance CNN method for offline handwritten chinese character recognition and visualization. Soft Comput. 2019;24:7977–87.
Li J, Mi Y, Li G, Ju Z. CNN-based facial expression recognition from annotated rgb-d images for human–robot interaction. Int J Humanoid Robot. 2019;16(04):504–5.
Kumar S, Singh SK. Occluded thermal face recognition using bag of CNN(BoCNN). IEEE Signal Process Lett. 2020;27:975–9.
Wang X. Deep learning in object recognition, detection, and segmentation. Found Trends Signal Process. 2016;8(4):217–382.
Article CAS Google Scholar
Boulent J, Foucher S, Théau J, St-Charles PL. Convolutional neural networks for the automatic identification of plant diseases. Front Plant Sci. 2019;10:941.
Article PubMed PubMed Central Google Scholar
Kumar S, Kaur R. Plant disease detection using image processing—a review. Int J Comput Appl. 2015;124(2):6–9.
Martineau M, Conte D, Raveaux R, Arnault I, Munier D, Venturini G. A survey on image-based insect classification. Pattern Recogn. 2016;65:273–84.
Jayme GAB, Luciano VK, Bernardo HV, Rodrigo VC, Katia LN, Claudia VG, et al. Annotated plant pathology databases for image-based detection and recognition of diseases. IEEE Latin Am Trans. 2018;16(6):1749–57.
Kaur S, Pandey S, Goel S. Plants disease identification and classification through leaf images: a survey. Arch Comput Methods Eng. 2018;26(4):1–24.
CAS Google Scholar
Shekhawat RS, Sinha A. Review of image processing approaches for detecting plant diseases. IET Image Process. 2020;14(8):1427–39.
Hinton GE, Salakhutdinov R. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7.
Liu W, Wang Z, Liu X, et al. A survey of deep neural network architectures and their applications. Neurocomputing. 2017;234:11–26.
Fergus R. Deep learning methods for vision. CVPR 2012 Tutorial; 2012.
Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798–828.
Article PubMed Google Scholar
Boureau YL, Le Roux N, Bach F, Ponce J, Lecun Y. [IEEE 2011 IEEE international conference on computer vision (ICCV)—Barcelona, Spain (2011.11.6–2011.11.13)] 2011 international conference on computer vision—ask the locals: multi-way local pooling for image recognition; 2011. p. 2651–8.
Zeiler MD, Fergus R. Stochastic pooling for regularization of deep convolutional neural networks. Eprint Arxiv. arXiv:1301.3557 . 2013.
TensorFlow. https://www.tensorflow.org/ .
Torch/PyTorch. https://pytorch.org/ .
Caffe. http://caffe.berkeleyvision.org/ .
Theano. http://deeplearning.net/software/theano/ .
Krizhenvshky A, Sutskever I, Hinton G. Imagenet classification with deep convolutional networks. In: Proceedings of the conference neural information processing systems (NIPS), Lake Tahoe, NV, USA, 3–8 December; 2012. p. 1097–105.
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions. In: Proceedings of the 2015 IEEE conference on computer vision and pattern recognition, Boston, MA, USA, 7–12 June; 2015. p. 1–9.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. arXiv:1409.1556 . 2014.
Xie S, Girshick R, Dollár P, Tu Z, He K. Aggregated residual transformations for deep neural networks. arXiv. arXiv:1611.05431 . 2017.
Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence. 2016.
Huang G, Lrj Z, Maaten LVD, et al. Densely connected convolutional networks. In: IEEE conference on computer vision and pattern recognition. 2017. p. 2261–9.
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H. MobileNets: efficient convolutional neural networks for mobile vision applications. arXiv. arXiv:1704.04861 . 2017.
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50 × fewer parameters and < 0.5 MB model size. arXiv. arXiv:1602.07360 . 2016.
Priyadharshini RA, Arivazhagan S, Arun M, Mirnalini A. Maize leaf disease classification using deep convolutional neural networks. Neural Comput Appl. 2019;31(12):8887–95.
Wen J, Shi Y, Zhou X, Xue Y. Crop disease classification on inadequate low-resolution target images. Sensors. 2020;20(16):4601.
Article PubMed Central Google Scholar
Thangaraj R, Anandamurugan S, Kaliappan VK. Automated tomato leaf disease classification using transfer learning-based deep convolution neural network. J Plant Dis Prot. 2020. https://doi.org/10.1007/s41348-020-00403-0 .
Atila M, Uar M, Akyol K, Uar E. Plant leaf disease classification using efficientnet deep learning model. Ecol Inform. 2021;61:101182.
Sabrol H, Kumar S. Recent studies of image and soft computing techniques for plant disease recognition and classification. Int J Comput Appl. 2015;126(1):44–55.
Yalcin H, Razavi S. Plant classification using convolutional neural networks. In: 2016 5th international conference on agro-geoinformatics (agro-geoinformatics). New York: IEEE; 2016.
Fuentes A, Lee J, Lee Y, Yoon S, Park DS. Anomaly detection of plant diseases and insects using convolutional neural networks. In: ELSEVIER conference ISEM 2017—The International Society for Ecological Modelling Global Conference, 2017. 2017.
Hasan MJ, Mahbub S, Alom MS, Nasim MA. Rice disease identification and classification by integrating support vector machine with deep convolutional neural network. In: 2019 1st international conference on advances in science, engineering and robotics technology (ICASERT). 2019.
Thenmozhi K, Reddy US. Crop pest classification based on deep convolutional neural network and transfer learning. Comput Electron Agric. 2019;164:104906.
Fang T, Chen P, Zhang J, Wang B. Crop leaf disease grade identification based on an improved convolutional neural network. J Electron Imaging. 2020;29(1):1.
Nagasubramanian K, Jones S, Singh AK, Sarkar S, Singh A, Ganapathysubramanian B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods. 2019;15(1):1–10.
Picon A, Seitz M, Alvarez-Gila A, Mohnke P, Echazarra J. Crop conditional convolutional neural networks for massive multi-crop plant disease classification over cell phone acquired images taken on real field conditions. Comput Electron Agric. 2019;167:105093.
Tianjiao C, Wei D, Juan Z, Chengjun X, Rujing W, Wancai L, et al. Intelligent identification system of disease and insect pests based on deep learning. China Plant Prot Guide. 2019;039(004):26–34.
Dechant C, Wiesner-Hanks T, Chen S, Stewart EL, Yosinski J, Gore MA, et al. Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology. 2017;107:1426–32.
Wiesner-Hanks T, Wu H, Stewart E, Dechant C, Nelson RJ. Millimeter-level plant disease detection from aerial photographs via deep learning and crowdsourced data. Front Plant Sci. 2019;10:1550.
Shougang R, Fuwei J, Xingjian G, Peishen Y, Wei X, Huanliang X. Deconvolution-guided tomato leaf disease identification and lesion segmentation model. J Agric Eng. 2020;36(12):186–95.
Fujita E, Kawasaki Y, Uga H, Kagiwada S, Iyatomi H. Basic investigation on a robust and practical plant diagnostic system. In: IEEE international conference on machine learning & applications. New York: IEEE; 2016.
Mohanty SP, Hughes DP, Salathé M. Using deep learning for image-based plant disease detection. Front Plant Sci. 2016;7:1419. https://doi.org/10.3389/fpls.2016.01419 .
Brahimi M, Arsenovic M, Laraba S, Sladojevic S, Boukhalfa K, Moussaoui A. Deep learning for plant diseases: detection and saliency map visualisation. In: Zhou J, Chen F, editors. Human and machine learning. Cham: Springer International Publishing; 2018. p. 93–117.
Chapter Google Scholar
Barbedo JG. Plant disease identification from individual lesions and spots using deep learning. Biosyst Eng. 2019;180:96–107.
Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39(6):1137–49.
Liu W, Anguelov D, Erhan D, Szegedy C, Berg AC. SSD: Single shot MultiBox detector. In: European conference on computer vision. Cham: Springer International Publishing; 2016.
Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real-time object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
Redmon J, Farhadi A. Yolo9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. p. 6517–25.
Redmon J, Farhadi A. Yolov3: an incremental improvement. arXiv preprint. arXiv:1804.02767 . 2018.
Fuentes A, Yoon S, Kim SC, Park DS. A robust deep-learning-based detector for real-time tomato plant diseases and pests detection. Sensors. 2017;17(9):2022.
Ozguven MM, Adem K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys A Statal Mech Appl. 2019;535(2019):122537.
Zhou G, Zhang W, Chen A, He M, Ma X. Rapid detection of rice disease based on FCM-KM and faster R-CNN fusion. IEEE Access. 2019;7:143190–206. https://doi.org/10.1109/ACCESS.2019.2943454 .
Xie X, Ma Y, Liu B, He J, Wang H. A deep-learning-based real-time detector for grape leaf diseases using improved convolutional neural networks. Front Plant Sci. 2020;11:751.
Singh D, Jain N, Jain P, Kayal P, Kumawat S, Batra N. Plantdoc: a dataset for visual plant disease detection. In: Proceedings of the 7th ACM IKDD CoDS and 25th COMAD. 2019.
Sun J, Yang Y, He X, Wu X. Northern maize leaf blight detection under complex field environment based on deep learning. IEEE Access. 2020;8:33679–88. https://doi.org/10.1109/ACCESS.2020.2973658 .
Bhatt PV, Sarangi S, Pappula S. Detection of diseases and pests on images captured in uncontrolled conditions from tea plantations. In: Proc. SPIE 11008, autonomous air and ground sensing systems for agricultural optimization and phenotyping IV; 2019. p. 1100808. https://doi.org/10.1117/12.2518868 .
Zhang B, Zhang M, Chen Y. Crop pest identification based on spatial pyramid pooling and deep convolution neural network. Trans Chin Soc Agric Eng. 2019;35(19):209–15.
Ramcharan A, McCloskey P, Baranowski K, Mbilinyi N, Mrisho L, Ndalahwa M, Legg J, Hughes D. A mobile-based deep learning model for cassava disease diagnosis. Front Plant Sci. 2019;10:272. https://doi.org/10.3389/fpls.2019.00272 .
Selvaraj G, Vergara A, Ruiz H, Safari N, Elayabalan S, Ocimati W, Blomme G. AI-powered banana diseases and pest detection. Plant Methods. 2019. https://doi.org/10.1186/s13007-019-0475-z .
Tian Y, Yang G, Wang Z, Li E, Liang Z. Detection of apple lesions in orchards based on deep learning methods of CycleGAN and YOLOV3-dense. J Sens. 2019. https://doi.org/10.1155/2019/7630926 .
Zheng Y, Kong J, Jin X, Wang X, Zuo M. CropDeep: the crop vision dataset for deep-learning-based classification and detection in precision agriculture. Sensors. 2019;19:1058. https://doi.org/10.3390/s19051058 .
Arsenovic M, Karanovic M, Sladojevic S, Anderla A, Stefanović D. Solving current limitations of deep learning based approaches for plant disease detection. Symmetry. 2019;11:21. https://doi.org/10.3390/sym11070939 .
Fuentes AF, Yoon S, Lee J, Park DS. High-performance deep neural network-based tomato plant diseases and pests diagnosis system with refinement filter bank. Front Plant Sci. 2018;9:1162. https://doi.org/10.3389/fpls.2018.01162 .
Jiang P, Chen Y, Liu B, He D, Liang C. Real-time detection of apple leaf diseases using deep learning approach based on improved convolutional neural networks. IEEE Access. 2019. https://doi.org/10.1109/ACCESS.2019.2914929 .
Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2015;39(4):640–51.
He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: 2017 IEEE international conference on computer vision (ICCV). New York: IEEE; 2017.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Berlin: Springer; 2015. p. 234–41. https://doi.org/10.1007/978-3-319-24574-4_28 .
Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2019;39(12):2481–95.
Wang Z, Zhang S. Segmentation of corn leaf disease based on fully convolution neural network. Acad J Comput Inf Sci. 2018;1:9–18.
Wang X, Wang Z, Zhang S. Segmenting crop disease leaf image by modified fully-convolutional networks. In: Huang DS, Bevilacqua V, Premaratne P, editors. Intelligent computing theories and application. ICIC 2019, vol. 11643. Lecture Notes in Computer Science. Cham: Springer; 2019. https://doi.org/10.1007/978-3-030-26763-6_62 .
Lin K, Gong L, Huang Y, Liu C, Pan J. Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front Plant Sci. 2019;10:155.
Kerkech M, Hafiane A, Canals R. Vine disease detection in UAV multispectral images using optimized image registration and deep learning segmentation approach. Comput Electron Agric. 2020;174:105446.
Stewart EL, Wiesner-Hanks T, Kaczmar N, Dechant C, Gore MA. Quantitative phenotyping of northern leaf blight in UAV images using deep learning. Remote Sens. 2019;11(19):2209.
Wang Q, Qi F, Sun M, Qu J, Xue J. Identification of tomato disease types and detection of infected areas based on deep convolutional neural networks and object detection techniques. Comput Intell Neurosci. 2019. https://doi.org/10.1155/2019/9142753 .
Hughes DP, Salathe M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. Comput Sci. 2015.
Shah JP, Prajapati HB, Dabhi VK. A survey on detection and classification of rice plant diseases. In: IEEE international conference on current trends in advanced computing. New York: IEEE; 2016.
Prajapati HB, Shah JP, Dabhi VK. Detection and classification of rice plant diseases. Intell Decis Technol. 2017;11(3):1–17.
Barbedo JGA, Koenigkan LV, Halfeld-Vieira BA, Costa RV, Nechet KL, Godoy CV, Junior ML, Patricio FR, Talamini V, Chitarra LG, Oliveira SAS. Annotated plant pathology databases for image-based detection and recognition of diseases. IEEE Latin Am Trans. 2018;16(6):1749–57.
Brahimi M, Arsenovic M, Laraba S, Sladojevic S, Boukhalfa K, Moussaoui A. Deep learning for plant diseases: detection and saliency map visualisation. In: Zhou J, Chen F, editors. Human and machine learning. Human–computer interaction series. Cham: Springer; 2018. https://doi.org/10.1007/978-3-319-90403-0_6 .
Tyr WH, Stewart EL, Nicholas K, Chad DC, Harvey W, Nelson RJ, et al. Image set for deep learning: field images of maize annotated with disease symptoms. BMC Res Notes. 2018;11(1):440.
Thapa R, Snavely N, Belongie S, Khan A. The plant pathology 2020 challenge dataset to classify foliar disease of apples. arXiv preprint. arXiv:2004.11958 . 2020.
Wu X, Zhan C, Lai YK, Cheng MM, Yang J. IP102: a large-scale benchmark dataset for insect pest recognition. In: 2019 IEEE/CVF conference on computer vision and pattern recognition (CVPR). New York: IEEE; 2019.
Huang M-L, Chuang TC. A database of eight common tomato pest images. Mendeley Data. 2020. https://doi.org/10.17632/s62zm6djd2.1 .
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Proceedings of the 2014 conference on advances in neural information processing systems 27. Montreal: Curran Associates, Inc.; 2014. p. 2672–80.
Pu Y, Gan Z, Henao R, et al. Variational autoencoder for deep learning of images, labels and captions [EB/OL]. 2016–09–28. arxiv:1609.08976 .
Oppenheim D, Shani G, Erlich O, Tsror L. Using deep learning for image-based potato tuber disease detection. Phytopathology. 2018;109(6):1083–7.
Too EC, Yujian L, Njuki S, Yingchun L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput Electron Agric. 2018;161:272–9.
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA. Using deep transfer learning for image-based plant disease identification. Comput Electron Agric. 2020;173:105393.
Zhang S, Huang W, Zhang C. Three-channel convolutional neural networks for vegetable leaf disease recognition. Cogn Syst Res. 2018;53:31–41. https://doi.org/10.1016/j.cogsys.2018.04.006 .
Liu B, Ding Z, Tian L, He D, Li S, Wang H. Grape leaf disease identification using improved deep convolutional neural networks. Front Plant Sci. 2020;11:1082. https://doi.org/10.3389/fpls.2020.01082 .
Karthik R, Hariharan M, Anand S, et al. Attention embedded residual CNN for disease detection in tomato leaves. Appl Soft Comput J. 2020;86:105933.
Guan W, Yu S, Jianxin W. Automatic image-based plant disease severity estimation using deep learning. Comput Intell Neurosci. 2017;2017:2917536.
Barbedo JGA. Factors influencing the use of deep learning for plant disease recognition. Biosyst Eng. 2018;172:84–91.
Barbedo JGA. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput Electron Agric. 2018;153:46–53.
Nawaz MA, Khan T, Mudassar R, Kausar M, Ahmad J. Plant disease detection using internet of thing (IOT). Int J Adv Comput Sci Appl. 2020. https://doi.org/10.14569/IJACSA.2020.0110162 .
Martinelli F, Scalenghe R, Davino S, Panno S, Scuderi G, Ruisi P, et al. Advanced methods of plant disease detection. A review. Agron Sustain Dev. 2015;35(1):1–25.
Liu J, Wang X. Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods. 2020;16:83.
Article CAS PubMed PubMed Central Google Scholar
Liu J, Wang X. Tomato diseases and pests detection based on improved Yolo V3 convolutional neural network. Front Plant Sci. 2020;11:898.
Kamal KC, Yin Z, Wu M, Wu Z. Depthwise separable convolution architectures for plant disease classification. Comput Electron Agric. 2019;165:104948.
Download references
Acknowledgements
Appreciations are given to the editors and reviewer of the Journal Plant Method.
This study was supported by the Facility Horticulture Laboratory of Universities in Shandong with Project Numbers 2019YY003, 2018YY016, 2018YY043 and 2018YY044; school level High-level Talents Project 2018RC002; Youth Fund Project of Philosophy and Social Sciences of Weifang College of Science and Technology with project numbers 2018WKRQZ008 and 2018WKRQZ008-3; Key research and development plan of Shandong Province with Project Number 2019RKA07012, 2019GNC106034 and 2020RKA07036; Research and Development Plan of Applied Technology in Shouguang with Project Number 2018JH12; 2018 innovation fund of Science and Technology Development centre of the China Ministry of Education with Project Number 2018A02013; 2019 basic capacity construction project of private colleges and universities in Shandong Province; and Weifang Science and Technology Development Programme with project numbers 2019GX081 and 2019GX082, Special project of Ideological and political education of Weifang University of science and technology (W19SZ70Z01).
Author information
Authors and affiliations.
Shandong Provincial University Laboratory for Protected Horticulture, Blockchain Laboratory of Agricultural Vegetables, Weifang University of Science and Technology, Weifang, 262700, Shandong, China
Jun Liu & Xuewei Wang
You can also search for this author in PubMed Google Scholar
Contributions
JL designed the research. JL and XW conducted the experiments and data analysis and wrote the manuscript. XW revised the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Correspondence to Xuewei Wang .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Liu, J., Wang, X. Plant diseases and pests detection based on deep learning: a review. Plant Methods 17 , 22 (2021). https://doi.org/10.1186/s13007-021-00722-9
Download citation
Received : 20 September 2020
Accepted : 13 February 2021
Published : 24 February 2021
DOI : https://doi.org/10.1186/s13007-021-00722-9
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Deep learning
- Plant diseases and pests
- Classification
- Object detection
- Segmentation
Plant Methods
ISSN: 1746-4811
- Submission enquiries: [email protected]
Information
- Author Services
Initiatives
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader.
All articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess .
Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications.
Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers.
Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal.
Original Submission Date Received: .
- Active Journals
- Find a Journal
- Proceedings Series
- For Authors
- For Reviewers
- For Editors
- For Librarians
- For Publishers
- For Societies
- For Conference Organizers
- Open Access Policy
- Institutional Open Access Program
- Special Issues Guidelines
- Editorial Process
- Research and Publication Ethics
- Article Processing Charges
- Testimonials
- Preprints.org
- SciProfiles
- Encyclopedia

Article Menu

- Subscribe SciFeed
- Recommended Articles
- Google Scholar
- on Google Scholar
- Table of Contents
Find support for a specific problem in the support section of our website.
Please let us know what you think of our products and services.
Visit our dedicated information section to learn more about MDPI.
JSmol Viewer
Lightweight multiscale cnn model for wheat disease detection.

1. Introduction
1.1. the significance of wheat disease detection, 1.2. disease identification in wheat based on machine learning and deep learning, 1.3. the advantages of lightweight models in wheat disease detection and the work of this article, 2. materials and methods, 2.1. image dataset, 2.2. dataset preprocessing, 2.3. proposed approach, 2.3.1. inception structure, 2.3.2. resnet model, 2.3.3. attentional mechanisms, 2.3.4. proposed model, 2.4. model optimization, 2.4.1. optimizer, 2.4.2. learning rate, 2.4.3. regularization, 2.5. model performance evaluation metrics, 3.1. comparison of effects of different optimizers, 3.2. exploring the impact of the inception module on the model, 3.3. effect of attentional mechanisms on the model, 3.4. comparison of the proposed model with the classical cnn model, 3.5. comparison of the proposed model with the classical lightweight model, 3.6. generalization ability test of the proposed model, 4. discussion, 5. conclusions, author contributions, institutional review board statement, informed consent statement, data availability statement, conflicts of interest.
- Sabenca, C.; Ribeiro, M.; Sousa, T.; Poeta, P.; Bagulho, A.S.; Igrejas, G. Wheat/Gluten-Related Disorders and Gluten-Free Diet Misconceptions: A Review. Foods 2021 , 10 , 1765. [ Google Scholar ] [ CrossRef ]
- Chai, Y.; Senay, S.; Horvath, D.; Pardey, P. Multi-peril pathogen risks to global wheat production: A probabilistic loss and investment assessment. Front. Plant Sci. 2022 , 13 , 1034600. [ Google Scholar ] [ CrossRef ]
- Biel, W.; Jaroszewska, A.; Stankowski, S.; Sobolewska, M.; Kępińska-Pacelik, J. Comparison of yield, chemical composition and farinograph properties of common and ancient wheat grains. Eur. Food Res. Technol. 2021 , 247 , 1525–1538. [ Google Scholar ] [ CrossRef ]
- Yao, F.; Li, Q.; Zeng, R.; Shi, S. Effects of different agricultural treatments on narrowing winter wheat yield gap and nitrogen use efficiency in China. J. Integr. Agric. 2021 , 20 , 383–394. [ Google Scholar ] [ CrossRef ]
- Kloppe, T.; Boshoff, W.; Pretorius, Z.; Lesch, D.; Akin, B.; Morgounov, A.; Shamanin, V.; Kuhnem, P.; Murphy, P.; Cowger, C. Virulence of Blumeria graminis f. sp. tritici in Brazil, South Africa, Turkey, Russia, and Australia. Adv. Breed. Wheat Dis. Resist. 2022 , 13 , 954958. [ Google Scholar ] [ CrossRef ]
- Mahum, R.; Munir, H.; Mughal, Z.-U.-N.; Awais, M.; Sher Khan, F.; Saqlain, M.; Mahamad, S.; Tlili, I. A novel framework for potato leaf disease detection using an efficient deep learning model. Hum. Ecol. Risk Assess. Int. J. 2023 , 29 , 303–326. [ Google Scholar ] [ CrossRef ]
- Zhang, J.; Pu, R.; Huang, W.; Yuan, L.; Luo, J.; Wang, J. Using in-situ hyperspectral data for detecting and discriminating yellow rust disease from nutrient stresses. Field Crops Res. 2012 , 134 , 165–174. [ Google Scholar ] [ CrossRef ]
- Zhang, D.; Lin, F.; Huang, Y.; Wang, X.; Zhang, L. Detection of Wheat Powdery Mildew by Differentiating Background Factors using Hyperspectral Imaging. Int. J. Agric. Biol. 2016 , 18 , 747–756. [ Google Scholar ] [ CrossRef ]
- Khan, I.H.; Liu, H.; Li, W.; Cao, A.; Wang, X.; Liu, H.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; et al. Early Detection of Powdery Mildew Disease and Accurate Quantification of Its Severity Using Hyperspectral Images in Wheat. Remote Sens. 2021 , 13 , 3612. [ Google Scholar ] [ CrossRef ]
- Wang, H.; Qin, F.; Liu, Q.; Ruan, L.; Wang, R.; Ma, Z.; Li, X.; Cheng, P.; Wang, H. Identification and Disease Index Inversion of Wheat Stripe Rust and Wheat Leaf Rust Based on Hyperspectral Data at Canopy Level. J. Spectrosc. 2015 , 2015 , 651810. [ Google Scholar ] [ CrossRef ]
- Bao, W.; Zhao, J.; Hu, G.; Zhang, D.; Huang, L.; Liang, D. Identification of wheat leaf diseases and their severity based on elliptical-maximum margin criterion metric learning. Sustain. Comput. Inform. Syst. 2021 , 30 , 100526. [ Google Scholar ] [ CrossRef ]
- Aboneh, T.; Rorissa, A.; Srinivasagan, R.; Gemechu, A. Computer Vision Framework for Wheat Disease Identification and Classification Using Jetson GPU Infrastructure. Technologies 2021 , 9 , 47. [ Google Scholar ] [ CrossRef ]
- Liu, X.; Zhou, S.; Chen, S.; Yi, Z.; Pan, H.; Yao, R. Buckwheat Disease Recognition Based on Convolution Neural Network. Appl. Sci. 2022 , 12 , 4795. [ Google Scholar ] [ CrossRef ]
- Jin, X.; Jie, L.; Wang, S.; Qi, H.; Li, S. Classifying Wheat Hyperspectral Pixels of Healthy Heads and Fusarium Head Blight Disease Using a Deep Neural Network in the Wild Field. Remote Sens. 2018 , 10 , 395. [ Google Scholar ] [ CrossRef ]
- Deng, J.; Lv, X.; Yang, L.; Zhao, B.; Zhou, C.; Yang, Z.; Jiang, J.; Ning, N.; Zhang, J.; Shi, J.; et al. Assessing Macro Disease Index of Wheat Stripe Rust Based on Segformer with Complex Background in the Field. Sensors 2022 , 22 , 5676. [ Google Scholar ] [ CrossRef ] [ PubMed ]
- Su, W.-H.; Zhang, J.; Yang, C.; Page, R.; Szinyei, T.; Hirsch, C.D.; Steffenson, B.J. Automatic Evaluation of Wheat Resistance to Fusarium Head Blight Using Dual Mask-RCNN Deep Learning Frameworks in Computer Vision. Remote Sens. 2020 , 13 , 26. [ Google Scholar ] [ CrossRef ]
- Shafi, U.; Mumtaz, R.; Qureshi, M.D.M.; Mahmood, Z.; Tanveer, S.K.; Haq, I.U.; Zaidi, S.M.H. Embedded AI for Wheat Yellow Rust Infection Type Classification. IEEE Access 2023 , 11 , 23726–23738. [ Google Scholar ] [ CrossRef ]
- Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Zhang, L.; Wen, S.; Zhang, H.; Zhang, Y.; Deng, Y. Detection of Helminthosporium Leaf Blotch Disease Based on UAV Imagery. Appl. Sci. 2019 , 9 , 558. [ Google Scholar ] [ CrossRef ]
- Pan, Q.; Gao, M.; Wu, P.; Yan, J.; Li, S. A Deep-Learning-Based Approach for Wheat Yellow Rust Disease Recognition from Unmanned Aerial Vehicle Images. Sensors 2021 , 21 , 6540. [ Google Scholar ] [ CrossRef ]
- Mi, Z.; Zhang, X.; Su, J.; Han, D.; Su, B. Wheat stripe rust grading by deep learning with attention mechanism and images from mobile devices. Front. Plant Sci. 2020 , 11 , 558126. [ Google Scholar ] [ CrossRef ]
- Bao, W.; Yang, X.; Liang, D.; Hu, G.; Yang, X. Lightweight convolutional neural network model for field wheat ear disease identification. Comput. Electron. Agric. 2021 , 189 , 106367. [ Google Scholar ] [ CrossRef ]
- Goyal, L.; Sharma, C.M.; Singh, A.; Singh, P.K. Leaf and spike wheat disease detection & classification using an improved deep convolutional architecture. Inform. Med. Unlocked 2021 , 25 , 100642. [ Google Scholar ]
- Zeng, W.; Li, M. Crop leaf disease recognition based on Self-Attention convolutional neural network. Comput. Electron. Agric. 2020 , 172 , 105341. [ Google Scholar ] [ CrossRef ]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [ Google Scholar ]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [ Google Scholar ]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [ Google Scholar ]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11534–11542. [ Google Scholar ]
- Gu, R.; Wang, G.; Song, T.; Huang, R.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. CA-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging 2020 , 40 , 699–711. [ Google Scholar ] [ CrossRef ]
- Rab Ratul, M.A.; Tavakol Elahi, M.; Yuan, K.; Lee, W. RAM-Net: A Residual Attention MobileNet to Detect COVID-19 Cases from Chest X-ray Images. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 195–200. [ Google Scholar ]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [ Google Scholar ]
- Liu, Y.; Shao, Z.; Teng, Y.; Hoffmann, N. NAM: Normalization-based Attention Module. arXiv 2021 , arXiv:2111.12419. [ Google Scholar ]
- Bottou, L. Stochastic gradient descent tricks. In Neural Networks: Tricks of the Trade , 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 421–436. [ Google Scholar ]
- Heo, B.; Chun, S.; Oh, S.J.; Han, D.; Yun, S.; Kim, G.; Uh, Y.; Ha, J.-W. Adamp: Slowing down the slowdown for momentum optimizers on scale-invariant weights. arXiv 2020 , arXiv:2006.08217. [ Google Scholar ]
- Mehta, S.; Paunwala, C.; Vaidya, B. CNN based traffic sign classification using adam optimizer. In Proceedings of the 2019 International Conference on Intelligent Computing and Control Systems (ICCS), Madurai, India, 15–17 May 2019; pp. 1293–1298. [ Google Scholar ]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015 , arXiv:1511.08060. [ Google Scholar ]
- Gokulnath, B. Identifying and classifying plant disease using resilient LF-CNN. Ecol. Inform. 2021 , 63 , 101283. [ Google Scholar ]
- Kukreja, V.; Kumar, D. Automatic classification of wheat rust diseases using deep convolutional neural networks. In Proceedings of the 2021 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 3–4 September 2021; pp. 1–6. [ Google Scholar ]
Click here to enlarge figure
| Wheat Types | Images | Training Images | Testing Images |
|---|---|---|---|
| healthy | 1086 | 869 | 217 |
| leaf rust | 1156 | 925 | 231 |
| powdery | 1380 | 1104 | 276 |
| wheat loose smut | 1342 | 1073 | 269 |
| root rot | 1096 | 877 | 219 |
| fusarium head blight | 1161 | 929 | 232 |
| tan spot | 1272 | 1018 | 254 |
| Network Layer | Inchannel, Outchannel, Kernel_Size, Stride, Padding | ||
|---|---|---|---|
| Image input | 224 × 224 × 3 | ||
| Inception-1 | branch1 | 3, 8, 1 × 1, 2, 0 | |
| branch2 | 3, 12, 1 × 1, 2, 0 | ||
| 12, 24, 3 × 3, 1, 1 | |||
| branch3 | —, —, 3 × 3, 1, 2 | ||
| 3, 8, 3 × 3, 1, 1 | |||
| branch4 | 3, 12, 1 × 1, 1, 0 | ||
| 12, 24, 3 × 3, 1, 1 | |||
| 24, 24, 3 × 3, 1, 1 | |||
| Filter concatenation | 112 × 112 × 64 | ||
| MaxPool | —, —, 3 × 3, 2, 1 | ||
| Residual-CE-1 | 64, 64, 3 × 3, 1, 1 | ||
| Residual-CE-1 | 64, 64, 3 × 3, 1, 1 | ||
| Residual-CE-2 | 64, 128, 3 × 3, 2, 1 | ||
| Residual-CE-1 | 128, 128, 3 × 3, 1, 1 | ||
| Inception-3 | branch1 | 128, 32, 1 × 1, 2, 0 | |
| branch2 | —, —, 3 × 3, 2, 1 | ||
| 128, 32, 1 × 1, 1, 0 | |||
| branch3 | 128, 64, 1 × 1, 2, 0 | ||
| 64, 64, 1 × 7, 1, [0, 3] | |||
| 64, 32, 7 × 1, 1, [3, 0] | |||
| branch4 | 128, 64, 1 × 1, 2, 0 | ||
| 64, 64, 1 × 7, 1, [0, 3] | |||
| 64, 64, 7 × 1, 1, [3, 0] | |||
| 64, 64, 1 × 7, 1, [0, 3] | |||
| 64, 32, 7 × 1, 1, [3, 0] | |||
| Filter concatenation | 14 × 14 × 128 | ||
| MaxPool | —, —, 3 × 3, 2, 1 | ||
| Residual-CE-2 | 128, 256, 3 × 3, 2, 1 | ||
| Residual-CE-1 | 256, 256, 3 × 3, 1, 1 | ||
| Inception-3 | branch1 | 256, 64, 1 × 1, 2, 0 | |
| branch2 | —, —, 3 × 3, 2, 1 | ||
| 256, 64, 1 × 1, 1, 0 | |||
| branch3 | 256, 128, 1 × 1, 2, 0 | ||
| 128, 96, 1 × 3, 1, [0, 1] | 128, 96, 3 × 1, 1, [1, 0] | ||
| branch4 | 256, 256, 1 × 1, 2, 0 | ||
| 256, 256, 3 × 1, 1, [1, 0] | |||
| 256, 256, 1 × 3, 1, [0, 1] | |||
| 256, 96, 1 × 3, 1, [0, 1] | 256, 96, 3 × 1, 1, [1, 0] | ||
| Filter concatenation | 2 × 2 × 512 | ||
| Avg_pool | 1 × 1 × 512 | ||
| Fc | 7 | ||
| Optimizer | Average Accuracy (%) |
|---|---|
| SGD | 75.97 ± 0.21 |
| RMSprop | 97.18 ± 0.15 |
| AdaGrad | 89.56 ± 0.13 |
| Adam | 98.64 ± 0.12 |
| Inception-1 | Inception-2 | Inception-3 | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Param (M) | FLOPs (G) |
|---|---|---|---|---|---|---|---|---|
| × | × | × | 95.40 | 95.53 | 95.52 | 95.53 | 3.06 | 0.42 |
| √ | × | × | 96.16 | 96.28 | 96.35 | 96.31 | 3.07 | 0.51 |
| √ | √ | × | 96.35 | 96.32 | 96.36 | 96.33 | 3.23 | 0.62 |
| √ | √ | √ | 98.76 | 98.77 | 98.81 | 98.79 | 4.24 | 0.84 |
| Methods | CBAM | ECA | SE | CA | NAM | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Param (M) | FLOPs (G) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IR | × | × | × | × | × | 96.05 | 96.13 | 96.17 | 96.15 | 4.22 | 0.83 |
| IRCBAM | √ | × | × | × | × | 96.46 | 96.54 | 96.55 | 96.55 | 4.24 | 0.84 |
| IRNAM | × | × | × | × | √ | 96.11 | 96.02 | 96.31 | 96.25 | 4.22 | 0.83 |
| IRSE | × | × | √ | × | × | 96.46 | 96.54 | 96.56 | 96.55 | 4.22 | 0.83 |
| IRCA | × | × | × | √ | × | 95.81 | 95.84 | 95.94 | 95.89 | 4.22 | 0.83 |
| IRECA | × | √ | × | × | × | 96.64 | 96.64 | 96.79 | 96.72 | 4.22 | 0.83 |
| IRCS | √ | × | √ | × | × | 97.23 | 97.32 | 97.25 | 97.27 | 4.24 | 0.84 |
| IRCC | √ | × | × | √ | × | 97.66 | 97.66 | 97.67 | 97.66 | 4.24 | 0.84 |
| IRCE | √ | √ | × | × | × | 98.76 | 98.77 | 98.81 | 98.79 | 4.24 | 0.84 |
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Param (M) | FLOPs (G) | Training Time (h) |
|---|---|---|---|---|---|---|---|
| AlexNet | 87.15 | 87.33 | 87.32 | 87.33 | 16.63 | 0.72 | 1.12 |
| VGG16 | 87.44 | 87.45 | 87.76 | 87.61 | 138.37 | 15.52 | 3.08 |
| ResNet34 | 95.05 | 95.09 | 95.07 | 95.08 | 11.69 | 3.61 | 1.20 |
| ResNet50 | 96.52 | 96.52 | 96.57 | 96.55 | 25.56 | 4.11 | 2.64 |
| ResNet101 | 95.68 | 95.68 | 95.65 | 95.66 | 44.55 | 7.82 | 2.62 |
| InceptionresnetV2 | 96.70 | 96.72 | 96.70 | 96.71 | 55.80 | 14.98 | 5.80 |
| IRCE | 98.76 | 98.77 | 98.81 | 98.79 | 4.24 | 0.84 | 1.34 |
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Param (M) | FLOPs (G) | Training Time (h) |
|---|---|---|---|---|---|---|---|
| MobileNetV1 | 94.41 | 94.42 | 94.47 | 94.49 | 2.59 | 0.33 | 1.52 |
| MobileNetV2 | 95.23 | 95.27 | 95.24 | 95.26 | 3.25 | 0.31 | 1.44 |
| MobileNetV3-Small | 95.34 | 95.44 | 95.42 | 95.43 | 2.54 | 0.06 | 1.36 |
| MobileNetV3-Large | 96.75 | 96.89 | 96.76 | 96.82 | 5.48 | 0.23 | 1.45 |
| EfficientNetb0 | 96.81 | 96.92 | 96.87 | 96.87 | 5.29 | 0.40 | 1.65 |
| IRCE | 98.76 | 98.77 | 98.81 | 98.79 | 4.24 | 0.84 | 1.34 |
| Dataset | Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|
| Plant-Village [ ] | LF-CNN [ ] | 98.93 | 95.61 | 97.16 | 96.65 |
| ResNet50 | 92.51 | 90.34 | 92.16 | 91.78 | |
| VGG16 | 94.13 | 92.10 | 93.18 | 92.35 | |
| MobileNetV2 | 97.81 | 97.42 | 97.93 | 97.65 | |
| IRCE | 99.74 | 99.71 | 99.66 | 99.68 | |
| CGIAR [ ] | ResNet34 | 92.11 | 92.52 | 92.74 | 92.65 |
| VGG19 | 94.40 | 94.52 | 95.31 | 94.82 | |
| EfficientNetb0 | 93.90 | 93.12 | 93.55 | 93.38 | |
| InceptionV3 | 95.72 | 95.32 | 95.71 | 95.42 | |
| IRCE | 96.70 | 96.70 | 96.70 | 96.70 | |
| Wheat Leaf Dataset | MobileNetV3_Large | 92.17 | 91.72 | 91.98 | 91.77 |
| ResNet34 | 89.92 | 89.94 | 91.01 | 90.91 | |
| EfficientNetb0 | 94.61 | 94.32 | 95.21 | 94.62 | |
| InceptionresnetV2 | 96.32 | 96.21 | 96.45 | 96.32 | |
| IRCE |
| Method | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Param (M) | FLOPs (G) | Training Time (h) |
|---|---|---|---|---|---|---|---|
| Method 1 | 95.28 | 95.52 | 95.38 | 95.45 | 13.39 | 2.04 | 1.52 |
| Method 2 | 95.23 | 95.27 | 95.24 | 95.26 | 4.02 | 0.73 | 1.03 |
| Method 3 | 98.76 | 98.77 | 98.81 | 98.79 | 4.24 | 0.84 | 1.34 |
| The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
Share and Cite
Fang, X.; Zhen, T.; Li, Z. Lightweight Multiscale CNN Model for Wheat Disease Detection. Appl. Sci. 2023 , 13 , 5801. https://doi.org/10.3390/app13095801
Fang X, Zhen T, Li Z. Lightweight Multiscale CNN Model for Wheat Disease Detection. Applied Sciences . 2023; 13(9):5801. https://doi.org/10.3390/app13095801
Fang, Xin, Tong Zhen, and Zhihui Li. 2023. "Lightweight Multiscale CNN Model for Wheat Disease Detection" Applied Sciences 13, no. 9: 5801. https://doi.org/10.3390/app13095801
Article Metrics
Article access statistics, further information, mdpi initiatives, follow mdpi.

Subscribe to receive issue release notifications and newsletters from MDPI journals

PLANT DISEASE DETECTION USING CONVOLUTIONAL NEURAL NETWORK
- Information Technology PSIT College of Engineering,Kanpur Uttar Pradesh, India.
- Computer Science and Engineering PSIT College of Engineering, KanpurUttar Pradesh, India.
- Cite This Article as
- Corresponding Author
When plants and crops are suffering from pests it affects the agricultural production of the country. Usually, farmers or experts observe the plants with eye for detection and identification of disease. But this method is often time processing, expensive and inaccurate. Automatic detection using image processing techniques provide fast and accurate results. This paper cares with a replacement approach to the development of disease recognition model, supported leaf image classification, by the utilization of deep convolutional networks. Advances in computer vision present a chance to expand and enhance the practice of precise plant protection and extend the market of computer vision applications within the field of precision agriculture. a completely unique way of training and therefore the methodology used facilitate a fast and straightforward system implementation in practice. All essential steps required for implementing this disease recognition model are fully described throughout the paper, starting from gathering images to make a database, assessed by agricultural experts, a deep learning framework to perform the deep CNN training. This method paper may be a new approach in detecting plant diseases using the deep convolutional neural network trained and finetuned to suit accurately to the database of a plants leaves that was gathered independently for diverse plant diseases. The advance and novelty of the developed model dwell its simplicity healthy leaves and background images are in line with other classes, enabling the model to distinguish between diseased leaves and healthy ones or from the environment by using CNN. Plants are the source of food on earth. Infections and diseases in plants are therefore a big threat, while the foremost common diagnosis is primarily performed by examining the plant body for the presence of visual symptoms [1]. As an alternative to the traditionally time-consuming process, different research works plan to find feasible approaches towards protecting plants. In recent years, growth in technology has engendered several alternatives to traditional arduous methods [2]. Deep learning techniques are very successful in image classification problems.
- Plant Disease Detection
- Machine Learning
- Image Processing
- Deep Learning
- Convolutional Neural Network
[ Prakanshu Srivastava, Kritika Mishra, Vibhav Awasthi, Vivek Kumar Sahu and Pawan Kumar Pal (2021); PLANT DISEASE DETECTION USING CONVOLUTIONAL NEURAL NETWORK Int. J. of Adv. Res. 9 (Jan). 691-698] (ISSN 2320-5407). www.journalijar.com
Article DOI: 10.21474/IJAR01/12346 DOI URL: http://dx.doi.org/10.21474/IJAR01/12346
Download Full Paper
Share this article.

Enhancing image classification using adaptive convolutional autoencoder-based snow avalanches algorithm
- Original Paper
- Published: 22 June 2024
Cite this article

- E. Dhiravidachelvi 1 ,
- T. Joshva Devadas 2 ,
- P. J. Sathish Kumar 3 &
- S. Senthil Pandi 4
21 Accesses
Explore all metrics
The disease that causes a large number of deaths annually across the world is brain cancer and it has become an important research topic in the field of medical image processing in recent times. There are various techniques for the detection of brain tumors (BT) but magnetic resonance imaging (MRI) diagnosing techniques show superior performance in the prognosis and examination of brain tumors in the early stages. The manual detection of brain tumors by radiologists leads to many limitations like errors and lack of detection accuracy. Hence, there is a need for computer-aided diagnostic techniques to help radiologists in detecting brain tumors accurately from the MRI images. To make this process more effective, the implementation of an automated technique is a preferred choice. In this paper, an effective detection and classification technique Adaptive convolutional Autoencoder-based Snow Avalanches (ACAE-SA) Algorithm is proposed. This algorithm comprises an Adaptive CNN component and an Autoencoder to detect and categorize BT from the MRI images. To mitigate the computational complexities in these components a Snow Avalanches algorithm is integrated into this work as an optimization technique. For the validation of the proposed architecture two MRI image datasets namely figshare and BraTS 2018 are used. The proposed technique proved its effectiveness in the detection and classification of brain tumors from the MRI images and outperformed the state-of-the-art techniques.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions

Similar content being viewed by others

Identification and Classification of Brain Tumor Using Convolutional Neural Network with Autoencoder Feature Selection

Political exponential deer hunting optimization-based deep learning for brain tumor classification using MRI

The Special Session Title ‘Intelligent Computing in Healthcare’ Dual Classification Framework for the Detection of Brain Tumor
Availability of data and material.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Mohan, P., Veerappampalayam Easwaramoorthy, S., Subramani, N., Subramanian, M., Meckanzi, S.: Handcrafted deep-feature-based brain tumor detection and classification using mri images. Electronics 11 (24), 4178 (2022)
Article Google Scholar
Saravanan, S., Kumar, V.V., Sarveshwaran, V., Indirajithu, A., Elangovan, D., Allayear, S.M.: Computational and mathematical methods in medicine glioma brain tumor detection and classification using convolutional neural network. Comput. Math. Methods Med. (2022)
Maqsood, S., Damaševičius, R., Maskeliūnas, R.: Multi-modal brain tumor detection using deep neural network and multiclass SVM. Medicina 58 (8), 1090 (2022)
Lamrani, D., Cherradi, B., El Gannour, O., Bouqentar, M.A., Bahatti, L.: Brain tumor detection using mri images and convolutional neural network. Int. J. Adv. Comput. Sci. Appl. 13 (7) (2022).
Saeedi, S., Rezayi, S., Keshavarz, H.R., Niakan Kalhori, S.: MRI-based brain tumor detection using convolutional deep learning methods and chosen machine learning techniques. BMC Med. Inf. Decision Making 23 (1), 16 (2023)
Uzun Ozsahin, D., Onakpojeruo, E.P., Uzun, B., Mustapha, M.T., Ozsahin, I.: Mathematical assessment of machine learning models used for brain tumor diagnosis. Diagnostics 13 (4), 618 (2023)
Aleid, A., Alhussaini, K., Alanazi, R., Altwaimi, M., Altwijri, O., Saad, A.S.: Artificial intelligence approach for early detection of brain tumors using MRI images. Appl. Sci. 13 (6), 3808 (2023)
Senan, E.M., Jadhav, M.E., Rassem, T.H., Aljaloud, A.S., Mohammed, B.A., Al-Mekhlafi, Z.G.: Early diagnosis of brain tumour mri images using hybrid techniques between deep and machine learning. Comput. Math. Methods Med. (2022)
Mehrotra, R., Ansari, M.A., Agrawal, R., Anand, R.S.: A transfer learning approach for AI-based classification of brain tumors. Mach. Learn. Appl. 2 , 100003 (2020)
Google Scholar
Jena, B., Saxena, S., Nayak, G.K., Balestrieri, A., Gupta, N., Khanna, N.N., Laird, J.R., Kalra, M.K., Fouda, M.M., Saba, L., Suri, J.S.: Brain tumor characterization using radiogenomics in artificial intelligence framework. Cancers 14 (16), 4052 (2022)
ZainEldin, H., Gamel, S.A., El-Kenawy, E.S.M., Alharbi, A.H., Khafaga, D.S., Ibrahim, A., Talaat, F.M.: Brain tumor detection and classification using deep learning and sine-cosine fitness grey wolf optimization. Bioengineering 10 (1), 18 (2022)
Amin, J., Sharif, M., Yasmin, M., Fernandes, S.L.: A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn. Lett. 139 , 118–127 (2020)
Yin, B., Wang, C., Abza, F.: New brain tumor classification method based on an improved version of whale optimization algorithm. Biomed. Signal Process. Control 56 , 101728 (2020)
Arif, M., Ajesh, F., Shamsudheen, S., Geman, O., Izdrui, D., Vicoveanu, D.: Brain tumor detection and classification by MRI using biologically inspired orthogonal wavelet transform and deep learning techniques. J. Healthcare Eng. (2022)
Abd El Kader, I., Xu, G., Shuai, Z., Saminu, S., Javaid, I., Ahmad, I.S., Kamhi, S.: Brain tumor detection and classification on MR images by a deep wavelet auto-encoder model. Diagnostics 11 (9), 1589 (2021)
Ait-Amou, M., Xia, K., Kamhi, S., Mouhafid, M.: A novel MRI diagnosis method for brain tumor classification based on CNN and Bayesian optimization. Healthcare 10 (3), 494 (2022)
Quang, S. L. Brats-2018. Kaggle. https://www.kaggle.com/datasets/sanglequang/brats2018 (2020 3).
Rathee, A. Figshare Brain Tumor Dataset. Kaggle. https://www.kaggle.com/datasets/ashkhagan/figshare-brain-tumor-dataset (2022, March 23).
Ojo, M.O., Zahid, A.: Improving deep learning classifiers performance via preprocessing and class imbalance approaches in a plant disease detection pipeline. Agronomy 13 (3), 887 (2023)
Jameel, S.M., Hashmani, M.A., Rehman, M., Budiman, A.: Adaptive CNN ensemble for complex multispectral image analysis. Complexity, pp. 1–21 (2020)
Ferreira, D., Silva, S., Abelha, A., Machado, J.: Recommendation system using autoencoders. Appl. Sci. 10 (16), 5510 (2020)
Golalipour, K., Nowdeh, S.A., Akbari, E., Hamidi, S.S., Ghasemi, D., Abdelaziz, A.Y., Yousef, A.: Snow avalanches algorithm (SAA): a new optimization algorithm for engineering applications. Alex. Eng. J. 83 , 257–285 (2023)
Alagarsamy, S., Kamatchi, K., Govindaraj, V., Zhang, Y.D., Thiyagarajan, A.: Multi-channeled MR brain image segmentation: A new automated approach combining BAT and clustering technique for better identification of heterogeneous tumors. Biocybern. Biomed. Eng. 39 (4), 1005–1035 (2019)
Nisha, A.V., Rajasekaran, M.P., Kottaimalai, R., Vishnuvarthanan, G., Arunprasath, T., Muneeswaran, V.: Hybrid D-OCapNet: automated multi-class Alzheimer’s disease classification in brain MRI using hybrid dense optimal capsule network. Int. J. Pattern Recogn. Artific. Intell. (2023).
Natarajan, S., Govindaraj, V., Murugan, P.R., Zhang, Y., Thiyagarajan, A.P., Uma, K.: Tumour region detection in MR brain images using MFCM based segmentation and Self Accommodative JAYA based optimization. In 2023 3rd International Conference on Advances in Computing, Communication, Embedded and Secure Systems (ACCESS) (pp. 340–345). IEEE (2023).
Alagarsamy, S., Govindaraj, V.: Automated brain tumor segmentation for MR brain images using artificial bee colony combined with interval type-II fuzzy technique. IEEE Trans. Ind. Inform. (2023).
Wang, S.H., Govindaraj, V.V., Górriz, J.M., Zhang, X., Zhang, Y.D.: Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inform. Fusion 67 , 208–229 (2021)
Download references
Acknowledgements
Not applicable
Not applicable.
Author information
Authors and affiliations.
Department of Electronics and Communication Engineering, Mohamed Sathak Engineering College, Kilakarai, Tamil Nadu, India
E. Dhiravidachelvi
School of Computer Science and Engineering, VIT University, Vellore, India
T. Joshva Devadas
Department of Computer Science and Engineering, Panimalar Engineering College, Chennai, Tamil Nadu, India
P. J. Sathish Kumar
Department of Computer Science and Engineering, Rajalakshmi Engineering College, Chennai, Tamil Nadu, India
S. Senthil Pandi
You can also search for this author in PubMed Google Scholar
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Dhiravidachelvi E, Joshva Devadas T, Sathish Kumar P.J, Senthil Pandi S. The first draft of the manuscript was written by Dhiravidachelvi E and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to E. Dhiravidachelvi .
Ethics declarations
Conflict of interest.
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Consent to participate
Consent for publication, additional information, publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Dhiravidachelvi, E., Devadas, T.J., Kumar, P.J.S. et al. Enhancing image classification using adaptive convolutional autoencoder-based snow avalanches algorithm. SIViP (2024). https://doi.org/10.1007/s11760-024-03357-0
Download citation
Received : 16 November 2023
Revised : 12 February 2024
Accepted : 21 February 2024
Published : 22 June 2024
DOI : https://doi.org/10.1007/s11760-024-03357-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Adaptive CNN
- AutoEncoder
- Brain tumor
- Medical image processing
- Snow avalanches algorithm
- Find a journal
- Publish with us
- Track your research

COMMENTS
Plant Disease Detection using CNN. Emma Harte. School of Computing. National College of Ireland. Mayor Street, IFSC, Dublin 1. Dublin, Ireland. [email protected]. Abstract — Plant ...
Among the 57 papers, 16 papers were selected for specific analysis based on the research object (plant disease) and research method (CNN). On this basis, the most recent research in 2022 is ...
A disease detection model by (Shelar et al., 2022) uses image processing with a CNN to aid in detecting plant illnesses. The study is driven by the fact that pests can majorly affect agricultural ...
Figure 3 depicts the number of research articles published on the automatic detection of plant leaf diseases using CNN from 2013 to 2022. The graph demonstrates that automatic disease detection research peaked in 2016, which confirmed the field's newness and freshness.
The proposed model using CNN was trained using images from plant village dataset and attained an accuracy of 94.87% in identifying the diseased plants with the help of image processing by OpenCV. Finally, the paper showcases the detailed analysis of the proposed scheme and results obtained by the model.
Early diagnosis methods have become a popular research topic for researchers. Plant disease detection using DL and image processing techniques have been among mostly addressed issues over the past decade. CNN has recently been successfully employed for many imaging applications, including plant disease detection (Keceli et al., 2022).
This method. paper may be a new approach in detecting plant diseases using the deep. convolutional neural network trained and finetuned to sui t accurately to. the database of a plant's leaves ...
The experiment uses the public data set new plant diseases dataset (NPDD) and compares it with ResNet50, DenseNet169, and EfficientNet. The experimental results show that the accuracy of FL-EfficientNet in identifying 10 diseases of 5 kinds of crops is 99.72%, which is better than the above comparison network.
The timely identification and early prevention of crop diseases are essential for improving production. In this paper, deep convolutional-neural-network (CNN) models are implemented to identify and diagnose diseases in plants from their leaves, since CNNs have achieved impressive results in the field of machine vision. Standard CNN models require a large number of parameters and higher ...
1. Development of an efficient model which could detect diseases on crops and plant leaves using CNN and deep learning techniques accurately. 2. Apply strategies to remove and avoid data imbalance as the data which is present is not balanced. 3.
The automatic detection of plant diseases using CNN was first performed by Sladojevic et al. 8 The proposed model was fine-tuned on CaffeNet, 9 that is, a variant of Alex Net, 10 using a self-collected dataset of 15 plant disease classes. The model attained the best average accuracy of 96.30%.
In the agricultural sector, the scourge of plant diseases casts a long shadow, leading to substantial financial losses and threatening both crop quality and quantity. With global food demands on the rise and the expansion of crop cultivation to meet these needs, the urgency for effective, scalable, and prompt plant disease detection methods cannot be overstated. Traditional approaches reliant ...
Plant diseases can have a significant influence on agricultural productivity, quality, and profitability. Early identification and precise diagnosis of plant diseases are essential for successful disease control, which may reduce the use of pesticides and minimize crop losses. In this study, a machine learning-based approach is developed to identify the diseases of various plants using leaf ...
This paper provides an overview of the current developments in the field of plant disease detection using ML and DL techniques. By covering research published between 2015 and 2022, it provides a comprehensive understanding of the state-of-the-art techniques and methodologies used in this field.
Hence in this paper, a novel hybrid model is proposed that reduces the dimensionality of input leaf image using CAE before classifying it using CNN for plant disease detection. The dimensionality reduction of plant leaf images before classification reduces the number of training parameters by a significant factor, which is the major finding of ...
Pepper leaf disease identification based on convolutional neural networks (CNNs) is one of the interesting research areas. However, most existing CNN-based pepper leaf disease detection models are suboptimal in terms of accuracy and computing performance. In particular, it is challenging to apply CN …
In this paper, deep convolutional-neural-network (CNN) models are implemented to. identify and diagnose diseases in plants from their leaves, since CNNs have achieved impressive. results in the ...
2. Plant Disease Detection by Well-Known DL Architectures. Many state-of-the-art DL models/architectures evolved after the introduction of AlexNet [] (as shown in Figure 3 and Table 1) for image detection, segmentation, and classification.This section presents the researches done by using famous DL architectures for the identification and classification of plants' diseases.
al in agriculture, as early detection and diagnosis of plant diseases can prevent crop losses. This paper presents an abstract of such a system that uses Convol. tional Neural Networks (CNN) to identify and classify plant disease based on images of leaves. The system includes pre-processing techniques for i.
A CNN based method for plant disease detection has been proposed here. Simulation study and analysis is done on sample images in terms of time complexity and the area of the infected region. ... A total of 15 cases have been fed to the model, out of which 12 cases are of diseased plant leaves namely, Bell Paper Bacterial Spot, Potato Early ...
Plant diseases and pests detection is a very important research content in the field of machine vision. It is a technology that uses machine vision equipment to acquire images to judge whether there are diseases and pests in the collected plant images [].At present, machine vision-based plant diseases and pests detection equipment has been initially applied in agriculture and has replaced the ...
Wheat disease detection is crucial for disease diagnosis, pesticide application optimization, disease control, and wheat yield and quality improvement. However, the detection of wheat diseases is difficult due to their various types. Detecting wheat diseases in complex fields is also challenging. Traditional models are difficult to apply to mobile devices because they have large parameters ...
98.59 percent in predicting plant disease. [11] S. Khirade and colleagues used digital image processing algorithms and BPNN - backpropagation neural networks to solve the problems of detection of plant diseases in 2015. Different techniques for identifying plant disease using photographs of leaves have been developed by the authors.
Approximately 35% of India's annual crop yield is lost due to plant diseases. Due to a lack of lab equipment and infrastructure, early diagnosis of plant diseases remains challenging. Categorization and detection of foliar diseases is a rapidly evolving research subject in which machine learning and neural computing concepts are employed to assist agricultural businesses. The lack of ...
This method paper may be a new approach in detecting plant diseases using the deep convolutional neural network trained and finetuned to suit accurately to the database of a plants leaves that was gathered independently for diverse plant diseases. The advance and novelty of the developed model dwell its simplicity healthy leaves and background ...
This paper proposes a method to classify leaf diseases. using the CNN model. Thus, in accord with to database. provided leaf diseases can be classified. Every plant disease can. be separately ...
The disease that causes a large number of deaths annually across the world is brain cancer and it has become an important research topic in the field of medical image processing in recent times. There are various techniques for the detection of brain tumors (BT) but magnetic resonance imaging (MRI) diagnosing techniques show superior performance in the prognosis and examination of brain tumors ...