

Introduction
Use of markers in screening for cancer, use of markers as diagnostic aids for cancer, use of markers in assessing prognosis, therapy-predictive markers in cancer, use of markers in surveillance following initial treatment for cancer, monitoring systemic therapy, acknowledgements, tumor markers in clinical practice: a review focusing on common solid cancers.
- Split-Screen
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Open the PDF for in another window
- Get Permissions
- Cite Icon Cite
- Search Site
Michael J. Duffy; Tumor Markers in Clinical Practice: A Review Focusing on Common Solid Cancers. Med Princ Pract 1 December 2012; 22 (1): 4–11. https://doi.org/10.1159/000338393
Download citation file:
- Ris (Zotero)
- Reference Manager
Tumor markers are playing an increasingly important role in cancer detection and management. These laboratory-based tests are potentially useful in screening for early malignancy, aiding cancer diagnosis, determining prognosis, surveillance following curative surgery for cancer, up front predicting drug response or resistance, and monitoring therapy in advanced disease. Clinically useful markers include fecal occult blood testing in screening for early colorectal cancer, carcinoembryonic antigen in the management of patients with colorectal cancer, both α-fetoprotein and human chorionic gonadotrophin in the management of patients with non-seminomatous germ cell tumors, CA 125 for monitoring therapy in patients with ovarian cancer, estrogen receptors for predicting response to hormone therapy in breast cancer, human epidermal growth factor receptor 2 for the identification of women with breast cancer likely to respond to trastuzumab (Herceptin) and KRAS mutational status for identifying patients with advanced colorectal cancer likely to benefit from treatment with the anti-epidermal growth factor receptor antibodies, cetuximab and panitumumab. Although widely used, the value of prostate-specific antigen screening in reducing mortality from prostate cancer is unclear.
Tumor markers may be defined as molecules which indicate the presence of cancer or provide information about the likely future behavior of a cancer (i.e., likelihood of progression or response to therapy) [ 1,2 ]. In asymptomatic subjects, tumor markers are potentially used in screening for early malignancy. In symptomatic patients, markers may help in the differential diagnosis of benign and malignant disease. Following diagnosis and surgical removal of a malignancy, markers may be used for assessing prognosis, postoperative surveillance, therapy prediction and monitoring response to systemic therapy [ 1,2 ].
Irrespective of its application, an ideal tumor marker should exhibit the following characteristics:
• possess a high positive and negative predictive value;
• have an inexpensive, simple, standardized and automated assay with clearly defined reference limits;
• be acceptable to subjects undergoing the test; and
• have its clinical value validated in a large prospective trial.
Suffice it to state, the ideal tumor marker does not currently exist. Despite this, several markers are now indispensable in the management of patients with cancer. The aims of this article are to review the most widely measured markers in clinical practice and summarize published guidelines for their use.
Screening involves the detection of early disease or a preclinical state in subjects without signs or symptoms of disease. Unlike disease diagnosis, screening is performed on individuals without any clinical sign of disease. Compared to established screening tests such as mammography for breast cancer, the Papanicolaou test for cervical cancer and colonoscopy for colorectal cancer (CRC), the use of tumor markers might be expected to have practical advantages in cancer screening [ 3 ]. These advantages include [ 3 ]:
• Markers can be measured in fluids such as blood and urine that can be obtained with minimal inconvenience to the individuals undergoing screening.
• For many markers, automated assays are available, allowing the processing of large numbers of samples in a relatively short period of time.
• Tests for markers provide quantitative results with objective endpoints.
• Assays for markers are relatively cheap compared to radiological, histological and endoscopy procedures.
Despite these advantages, markers have several limitations as cancer screening tests. In particular, lack of sensitivity for early invasive disease or premalignant lesions and lack of specificity for malignancy limit the use of existing markers in screening asymptomatic subjects for early malignancy [ 1,3 ]. The dual problem of limited sensitivity and specificity, especially when combined with the low prevalence of most cancers in the community, means that markers, if used alone, have low positive predictive values in screening asymptomatic populations. Indeed, it is the low prevalence of cancer in the general population that prohibits most biomarkers from being used in screening for cancer [ 1,3 ]. Despite this, a number of markers have either undergone or are presently undergoing evaluation for cancer screening (table 1 ). Some of the most widely investigated markers in cancer screening are now discussed.
Biomarkers that have undergone or are currently undergoing evaluation in screening asymptomatic subjects for cancer
Fecal Occult Blood Testing in Screening for CRC
One of the best validated screening markers is the use of fecal occult blood testing (FOBT) in screening for CRC. Indeed, 4 large randomized prospective trials carried out in Europe and North America have all shown that screening apparently healthy subjects over 50 years of age using FOBT significantly reduced mortality from CRC [ 4 ]. Combined results from the 4 trials showed that the screening resulted in a 16% reduction in the relative risk of CRC mortality [ 4 ]. Following adjustment for those subjects that failed to attend screening, mortality reduction increased to 25%.
Screening healthy individuals aged ≥50 years using FOBT is now recommended, especially in Europe [ 5,6 ]. Although the 4 prospective randomized trials mentioned above all used a guaiac-based FOBT, both the European Group of Tumor Markers (EGTM) and a European Union expert panel currently recommend the use of a fecal immunochemical test in screening for CRC [ 5,6 ]. As previously reviewed, the use of fecal immunochemical tests have several advantages vis-à-vis FOBTs, including greater analytical sensitivity and specificity, improved clinical performance and potential for automation [ 5 ].
Prostate-Specific Antigen in Screening for Prostate Cancer
In contrast to FOBT in screening for CRC, the clinical value of prostate-specific antigen (PSA) in screening for prostate cancer is less clear [ 7 ]. In 2009, results from 2 large randomized prospective trials evaluating the screening potential of this marker were published. The first of these trials was carried out in the USA and involved 76,693 men that were randomized to either annual screening or regular care [ 8 ]. Following analysis at 7–10 years of follow-up, death rates were similar in the 2 groups. A limitation of this study was that approximately 50% of men in the control group underwent screening during the study. This trial could thus be regarded as a comparison between a heavily screened group and a less heavily screened group rather than a true randomized trial [ 8 ]. A further limitation was that approximately 40% of the men participating had a PSA test prior to the start of the trial. These men were thus less likely to be diagnosed with prostate cancer during the trial proper.
In a somewhat similar but larger trial carried out in Europe, 162,243 men were randomly allocated to PSA screening or to a control group not subjected to screening [ 9 ]. After a median follow-up of 9 years, death rates from prostate cancer were found to be 20% lower in the screened compared to the control group. This apparent benefit, however, resulted in major overdetection and overtreatment. Thus, according to the paper’s authors, 1,410 men would have to be screened and 48 additional cases of prostate cancer would have to undergo treatment to prevent 1 death from prostate cancer [ 9 ].
With these conflicting findings it is not surprising that guidelines published by expert panels vary with respect to PSA screening for prostate cancer, i.e., while some groups recommend screening, others are opposed to the practice [ 7,10 ]. Although expert panels disagree on whether or not to recommend PSA screening, most recommend that prior to undergoing screening for prostate cancer, men should be informed of the risks and benefits of early disease detection [ 7 ]. Only after receiving such information should PSA screening be initiated.
CA 125 in Screening for Ovarian Cancer
For the past decade or so, 2 large prospective randomized trials have been evaluating the role of CA 125 and transvaginal ultrasound in screening for ovarian cancer [ 11,12 ]. One of these trials recently reported its findings [ 13 ]. In this trial carried out in the USA, 78,216 women aged 55–74 years were randomized to undergo either annual screening with CA 125 for 6 years and transvaginal ultrasound for 4 years or standard care. After 13 years of follow-up, death rates were similar in the screened and control groups. Clearly, in this trial, screening with CA 125 and transvaginal ultrasound did not reduce ovarian cancer mortality [ 13 ]. According to the EGTM guidelines, CA 125 either alone or in combination with other modalities cannot be recommended in screening for ovarian cancer in asymptomatic women outside the context of a randomized controlled trial [ 14 ].
α-Fetoprotein in Hepatocellular Cancer
Subjects at increased risk of developing hepatocellular cancer (HCC) include those with cirrhosis due to infection with hepatitis B virus or hepatitis C virus, genetic hemochromatosis or biliary cirrhosis. Guidelines published by expert panels differ in their recommendations regarding the use of α-fetoprotein (AFP) in screening for HCC. Thus, the National Academy of Clinical Biochemistry USA (NACB) [ 15 ] state that, ‘AFP should be measured and abdominal ultrasound performed at six-monthly intervals in patients at high risk of HCC, especially in those with hepatitis B and hepatitis C-related liver cirrhosis. AFP concentrations that are >20 µg/l and showing consistent increases in concentration should prompt further investigation even if ultrasound is negative.’ On the other hand, the American Association for the Study of Liver Disease recommend that surveillance of high-risk subjects should be performed only with ultrasound. According to this organization, AFP should only be used when ultrasound is not available. This latter recommendation is based on the limited use of AFP in detecting early HCC.
As discussed above with screening, limited sensitivity for small or early cancers and lack of tumor specificity preclude the use of serum markers for the primary diagnosis of cancer. In a limited number of situations, however, markers may aid in the differential of benign and malignant disease. Thus, CA 125 is used as an adjunct in differentiating between benign and malignant pelvic masses in postmenopausal women [ 14 ]. The EGTM recommend that CA 125 should be measured in postmenopausal women presenting with such masses. According to this expert panel, patients with elevated levels (e.g. >35 U/l) should be considered for referral to a surgeon specialized in gynecological oncology surgery [ 14 ]. This recommendation was based on studies showing superior outcome in patients with ovarian cancer if treated by a gynecological oncologist rather than by a general surgeon [ 16 ].
Another marker that may be helpful in cancer diagnosis is AFP in the detection of HCC. According to the American Association for the Study of Liver Disease, finding a hepatic mass of >2 cm in diameter in a patient with a cirrhotic liver is highly suspicious of HCC. If AFP is >200 µg/l and the radiological appearance is suggestive of HCC, the likelihood is that the lesion is HCC and biopsy is not essential [ 17 ]. Similar recommendations have been published by the NACB [ 15 ]. AFP, however, is of limited value in aiding the diagnosis of lesions <2 cm in diameter.
Prognostic markers provide information on the likely outcome following diagnosis of a disease. Such markers may help avoid undertreatment of patients with aggressive disease and overtreatment of those with indolent disease. Prognostic markers are most important in cancers that vary widely in their outcome such as prostate and breast cancer. In these cancers, prognostic markers may help identify those patients with aggressive disease that could benefit from additional therapies and simultaneously select those patients who may not require additional therapy.
Prognostic markers are most widely used in breast cancer, especially in the subset with lymph node-negative disease. In this subgroup of patients with breast cancer, prognostic markers help identify patients who may be spared the toxicity and side effects of adjuvant chemotherapy. The 2 best validated prognostic markers for breast cancer are the Oncotype DX test and uPA/PAI1. The Oncotype DX test measures the expression of 16 cancer-associated and 5 control genes by RT-PCR on RNA isolated from paraffin-embedded breast cancer tissue [ 18 ]. This test is currently available from a commercial laboratory in the USA. Although the Oncotype DX test is recommended by several expert panels including the American Society of Clinical Oncology (ASCO), NACB and EGTM [ 10,19,20 ], the test has not yet been validated in a large prospective randomized trial. Such a study, however, is ongoing as part of the TAILORx trial.
uPA and PAI-1 are the best validated prognostic markers in breast cancer, especially for patients with lymph node-negative disease [ 21 ]. uPA is a protease that mediates invasion and metastasis. Although PAI-1 normally functions as an endogenous inhibitor of uPA activity, at the high concentrations frequently present in tumors it appears, like uPA, to also play a role in cancer spread [ 21 ]. Unlike any other marker in breast cancer, the prognostic impact of uPA and PAI-1 has been validated in both a multicenter randomized prospective trial and a pooled analysis of raw data from several small retrospective and a prospective study [ 22,23 ], i.e., in 2 level I evidence studies.
As with Oncotype DX, measurement of uPA/PAI-1 is recommended by a number of expert panels [ 10,19,20 ]. According to ASCO, ‘uPA and PAI-1 may be used for the determination of prognosis in patients with newly diagnosed, node negative breast cancer. Low levels of both markers are associated with a sufficiently low risk of recurrence, especially in hormone receptor-positive women who will receive adjuvant endocrine therapy, that chemotherapy will only contribute minimal additional benefit. Furthermore, CMF-based adjuvant chemotherapy provides substantial benefit, compared with observation alone, in patients with high risk of recurrence as determined by high levels of uPA and PAI-1’ [ 19 ]. Other widely studied cancer prognostic markers are listed in table 2 .
Markers that may be used for determining prognosis in different cancers
As mentioned above, therapy-predictive markers are factors that prospectively identify likely response or resistance to a specific treatment. Predictive markers are important in cancer patient management as patients with the same histological type of malignancy respond very differently to a specific drug. Thus, response rates for patients with different types of advanced cancer to currently available systemic treatments vary from about 10 to >90% [ 24 ]. Many of the newer biological therapies, in particular, have efficacy in only a minority of patients. This finding, when combined with the high costs of these drugs [ 25 ], illustrates the importance of having accurate predictive markers.
As well as assessing efficacy, predictive markers may also be able to identify optimum drug dose and predict toxicity. Thus, measurement of predictive markers can increase drug efficacy and result in decreased toxicity. This, in turn, should reduce overall health care costs and lead to an enhanced quality of life for patients [ 26 ].
As with prognostic markers, predictive markers are most developed for breast cancer. Thus, in breast cancer, measurement of estrogen receptors and progesterone receptors is universally used to identify patients for treatment with hormone therapy (tamoxifen or aromatase inhibitor), while the assay of human epidermal growth factor receptor 2 is routinely used to select patients for treatment with trastuzumab (Herceptin) or lapatinib [ 27 ]. Newly introduced predictive markers include mutant KRAS status for identifying responsiveness to anti-epidermal growth factor receptor antibodies (cetuximab and panitumumab) in advanced CRC and epidermal growth factor receptor mutation status for selecting patients with advanced non-small cell lung cancer for treatment with anti-epidermal growth factor receptor kinases (gefitinib and erlotinib) [ 27 ].
One of the most frequent uses of tumor markers at present is in the postoperative follow-up of patients following a diagnosis of malignancy (table 3 ). In several situations, serial levels of cancer markers can predict the presence of early recurrent/metastatic disease, i.e., provide a lead time over clinical or radiological findings. It is assumed that the early detection of recurrent/metastatic disease followed by the initiation of treatment enhances the chance of cure or results in an improved survival [ 2 ]. For some cancer types, however, the evidence currently available does not support this widely held assumption. Indeed, the value of makers in postoperative surveillance may vary from cancer to cancer. Below, I discuss the usefulness of markers in surveillance following the diagnosis of different cancers.
Serum markers that may be used in postoperative surveillance and monitoring therapy in different cancers
Carcinoembryonic Antigen in CRC
Multiple studies have shown that following curative surgery for CRC, patients undergoing an intensive surveillance regime that included regular carcinoembryonic antigen (CEA) measurements had a significantly better outcome than those undergoing surveillance without CEA testing [ 28,29 ]. Most expert panels in Europe and the USA therefore recommend serial measurements of CEA following curative surgery for CRC [ 10,30,31,32 ]. According to the EGTM, ‘CRC patients with stage II and III (Dukes’ B and C) CRC that may be candidates for either liver resection or systemic treatment in the event of recurrence in that organ, should have CEA measured every 2 to 3 months for at least 3 years after diagnosis’ [ 30,31 ]. Although serial measurements of CEA are widely recommended as part of a surveillance regime, agreement is lacking as to the extent of concentration change that constitutes a clinically significant increase in marker levels. According to the EGTM group [ 30 ], ‘a significant increase in CEA levels occurs if the elevation is at least 30% over that of the previous concentration’. This organization also states that prior to initiating therapy, any increase must be confirmed by a second sample taken within approximately 1 month. If the second sample is also increased, the patient should undergo further investigations such as imaging [ 30 ].
AFP and Human Chorionic Gonadotrophin in Patients with Germ Cell Tumors
Two main histological types of germ cell tumors exist, seminoma and non-seminoma. The use of AFP and human chorionic gonadotrophin (HCG) in monitoring patients with the non-seminomatous type germ cell tumors of the testis is often regarded as approximating the ideal use of tumor markers [ 2 ]. This is because these 2 markers are sensitive indicators of germ cell disease status, i.e., whether disease is stable, progressing or regressing. A further reason why these markers are particularly helpful in patients with germ cell tumors is that these malignancies are highly chemosensitive. Indeed, it is now widely accepted that following orchidectomy for non-seminomatous testicular germ cell tumors, increasing AFP or HCG levels in the absence of radiological or clinical evidence of disease suggests active disease and may provide sufficient reassurance to initiate treatment, provided likely causes of false-positive marker levels can be eliminated [ 2 ].
According to the ASCO guidelines, AFP and HCG should be assayed during surveillance following definite therapy for non-seminomatous germ cell tumors, regardless of stage [ 33 ]. These measurements may be carried out every 1–2 months in the first year, every 2–4 months in the second year, every 3–6 months in the third and fourth years, every 6 months in the fifth year and annually thereafter. Surveillance should be continued for at least 10 years after therapy is completed. For monitoring patients with pure seminoma, HCG and/or LDH are generally recommended [ 33 ].
CA 125 in Patients with Ovarian Cancer
The clinical value of serial determination of CA 125 in post-therapy monitoring of patients with a history of ovarian cancer is unclear. Although regular measurement of the marker may detect early recurrences with a median lead time of 4–5 months [ 34 ], a recently completed prospective randomized trial found no survival benefit from starting early treatment based on a rising serum CA 125 level [ 35 ]. This trial involved 1,442 women previously diagnosed with ovarian cancer but in clinical remission. CA 125 levels were measured every 3 months, but the results were not made available to patients or their doctors. The women were randomized when their marker concentrations reached twice the upper limit of the normal range, to receive treatment immediately or to continue with blinded CA 125 determinations. In this latter situation, women underwent treatment only when there was clinical evidence of recurrence. Despite the earlier introduction of second-line chemotherapy, no significant difference in overall survival was found in the 2 groups. This negative finding may relate, at least in part, to the lack of effective therapy for recurrent ovarian cancer.
Uncertainties therefore exist with respect to the value of CA 125 measurement and timing of treatment for relapsed ovarian cancer. Although the NACB panel currently recommends serial measurement of CA 125 following surgery and initial systemic therapy for ovarian cancer [ 10 ], the EGTM panel is opposed to this practice [ 14 ]. With such conflicting recommendations, a practical way forward may be to take the patients’ wishes into consideration.
PSA in Prostate Cancer
Irrespective of whether men with diagnosed prostate cancer undergo active treatment (e.g. with radical prostatectomy, radiotherapy or brachytherapy) or active surveillance, regular monitoring with PSA is now commonly carried out [ 36 ]. Following successful radical prostatectomy, PSA levels should decline to undetectable levels. A subsequent increase to ≥0.2 µg/l is defined as biochemical recurrence [ 37 ]. Generally, decreases in PSA levels following radiation therapy are less than those following radical prostatectomy. A rise in PSA levels of ≥2 µg/l over and above the nadir value has been proposed as a definition of radiation therapy failure [ 38 ]. It is still unclear whether the introduction of salvage therapy based on these definitions of PSA recurrence enhances patient outcome or quality of life.
CA 15-3 in Breast Cancer
The most widely used marker in surveillance following a diagnosis of breast cancer is CA 15-3 [ 39 ]. Although widely used in some countries for this purpose, it is unclear whether serial measurement improves patient outcome. Consequently, guidelines vary with respect to their recommendations on the use of CA 15-3 in monitoring asymptomatic women following a diagnosis of breast cancer. While expert panels such as ASCO recommend against routing use of CA 15-3 in surveillance in the surveillance of breast cancer patients [ 19 ], EGTM endorses its measurement in this setting [ 20 ]. According to the NACB guidelines [ 10 ], ‘CA 15-3 should not be routinely used for the early detection of recurrences/metastases in patients with diagnosed breast cancer. However, as some patients, as well as some doctors, may wish to have these measurements, the ultimate decision on whether or not to use CA 15-3 must be taken by the doctor in consultation with the patient.’
Another frequent use of tumor markers is in monitoring patients with advanced cancer receiving systemic therapy (table 3 ) [ 1,2,10 ]. The markers used in monitoring therapy in a specific malignancy are the same as those measured in postoperative surveillance (see above). Generally, decreasing levels of markers following the initiation of therapy correlates with tumor regression and increasing levels predict progressive disease. Tumor markers, however, should not be used alone in assessing response to therapy.
A caveat in the use of markers in monitoring therapy in patients with advanced malignancy is the possible occurrence of transient increases or spikes within the first few weeks of administering therapy. These transient increases appear to be due to tumor cell necrosis or apoptosis in response to the initial treatment with chemotherapy. Such transient increases have not yet been reported with biological therapies such as therapeutic antibodies (e.g. Herceptin, cetuximab or panitumumab).
From above, it is clear that certain tumor markers are mandatory in the management of patients with certain types of cancer. Indeed, in some situations, markers can be used as the sole criterion for clinical decision making. This applies particularly for therapy-predictive markers such as estrogen receptor and human epidermal growth factor receptor 2 in breast cancer. Another good example is the use of HCG and AFP in therapy decision making in patients with diagnosed non-seminomatous germ cell tumors. In other situations, however, the value of markers is less clear, for example, the role of PSA in screening for prostate cancer, assay of CA 125 following surgery and chemotherapy for ovarian cancer and assay of CA 15-3 in the surveillance of patients following the diagnosis of breast cancer. Hopefully, future studies will provide definite answers on the use of these markers, in the near future. New procedures for measuring tumor markers in the future are likely to focus on gene expression microarray, proteomics and detection of circulating tumor cells.
The authors wish to thank Science Foundation Ireland, Strategic Research Cluster Award (08/SRC/B1410) to Molecular Therapeutics for Cancer Ireland.

Email alerts
Citing articles via, suggested reading.
- Online ISSN 1423-0151
- Print ISSN 1011-7571
INFORMATION
- Contact & Support
- Information & Downloads
- Rights & Permissions
- Terms & Conditions
- Catalogue & Pricing
- Policies & Information
- People & Organization
- Stay Up-to-Date
- Regional Offices
- Community Voice
SERVICES FOR
- Researchers
- Healthcare Professionals
- Patients & Supporters
- Health Sciences Industry
- Medical Societies
- Agents & Booksellers
Karger International
- S. Karger AG
- P.O Box, CH-4009 Basel (Switzerland)
- Allschwilerstrasse 10, CH-4055 Basel
- Tel: +41 61 306 11 11
- Fax: +41 61 306 12 34
- Contact: Front Office
- Experience Blog
- Privacy Policy
- Terms of Use
This Feature Is Available To Subscribers Only
Sign In or Create an Account
Tumor markers in clinical practice: a review focusing on common solid cancers
Affiliation.
- 1 Department of Pathology and Laboratory Medicine, St Vincent's University Hospital, Dublin, Ireland. [email protected]
- PMID: 22584792
- PMCID: PMC5586699
- DOI: 10.1159/000338393
Tumor markers are playing an increasingly important role in cancer detection and management. These laboratory-based tests are potentially useful in screening for early malignancy, aiding cancer diagnosis, determining prognosis, surveillance following curative surgery for cancer, up front predicting drug response or resistance, and monitoring therapy in advanced disease. Clinically useful markers include fecal occult blood testing in screening for early colorectal cancer, carcinoembryonic antigen in the management of patients with colorectal cancer, both α-fetoprotein and human chorionic gonadotrophin in the management of patients with non-seminomatous germ cell tumors, CA 125 for monitoring therapy in patients with ovarian cancer, estrogen receptors for predicting response to hormone therapy in breast cancer, human epidermal growth factor receptor 2 for the identification of women with breast cancer likely to respond to trastuzumab (Herceptin) and KRAS mutational status for identifying patients with advanced colorectal cancer likely to benefit from treatment with the anti-epidermal growth factor receptor antibodies, cetuximab and panitumumab. Although widely used, the value of prostate-specific antigen screening in reducing mortality from prostate cancer is unclear.
Copyright © 2012 S. Karger AG, Basel.
Publication types
- Research Support, Non-U.S. Gov't
- Biomarkers, Tumor / blood*
- CA-125 Antigen / blood
- Carcinoembryonic Antigen / blood
- Chorionic Gonadotropin / blood
- Early Detection of Cancer / methods*
- Environmental Monitoring
- Mucin-1 / blood
- Neoplasms / diagnosis*
- Occult Blood
- Prostate-Specific Antigen / blood
- alpha-Fetoproteins / analysis
- Biomarkers, Tumor
- CA-125 Antigen
- Carcinoembryonic Antigen
- Chorionic Gonadotropin
- alpha-Fetoproteins
- Prostate-Specific Antigen
Advertisement
Cancer Biomarkers: Status and Its Future Direction
- Review Article
- Published: 20 February 2023
- Volume 85 , pages 1323–1335, ( 2023 )
Cite this article

- Tongbram Malemnganbi Chanu 1 ,
- Lakhon Kma 2 &
- R. N. Sharan ORCID: orcid.org/0000-0002-7120-8023 1
277 Accesses
2 Citations
Explore all metrics
Biomarkers are helpful for disease diagnosis, monitoring disease progression, predicting disease recurrence, treatment monitoring, and efficacy, especially in the domain of cancer management and therapeutics. Clinical cancer biomarker utilization is progressively increasing with increase in and better access to healthcare by the growing populations worldwide. Early cancer detection and therapeutics enhancement remain at the core to increase our cancer management and control abilities. A biomarker can help detect cancer at an early stage as well as aid the clinicians in individualization of therapeutics enhancing the clinical efficacy of cancer therapy. Over the years, several biomarkers have been established and are in regular use. Nonetheless, discovering new cancer biomarkers, which are more sensitive, specific, and clinically convenient, remains important. The current review critically analyzes the existing and established cancer biomarkers as well as futuristic biomarkers for clinicians, surgeons, oncologists, and researchers. A tabular summary at the end of the article and compilation of up to date literature in the domain should be useful ready reckoners for clinicians, oncologists, and researchers alike.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others

Overview of Oncology Biomarkers

Cancer Biomarkers Discovery and Validation: State of the Art, Problems and Future Perspectives
Abbreviations.
- Average cancer therapy
Alpha-fetoprotein
ADP-ribose polymer adduct
Breast cancer–associated genes
Cancer antigen 125
Cancer antigen 15–3
Cancer antigen 19–9 or carbohydrate antigen 19–9
Cancer antigen 27–29
Cancer biomarkers
Clinical cancer biomarkers
Cyclin-dependent kinase inhibitor 1A
Carcinoembryonic antigen
Cancer therapy or cancer therapeutics
Experimental cancer biomarkers
Human chorionic gonadotrophin
Human epididymis protein 4
Human epidermal growth factor receptor 2
Heat-shock protein
Multivariate index assay
Mammalian target of rapamycin
Nuclear factor 1-C
Ovarian Malignancy Algorithm
Personalized and precise cancer therapy
Prostate-specific antigen
Risk of Ovarian Malignancy Algorithm
Thyroglobulin
Vascular endothelial growth factor
Lassere MN (2008) The Biomarker-Surrogacy Evaluation Schema: a review of the biomarker-surrogate literature and a proposal for a criterion-based, quantitative, multidimensional hierarchical levels of evidence schema for evaluating the status of biomarkers as surrogate endpoints. Stat Methods Med Res 17:303–340
Article PubMed Google Scholar
World Health Organization & International Programme on Chemical Safety (1993) Biomarkers and risk assessment: concepts and principles / published under the joint sponsorship of the United Nations environment Programme, the International Labour Organisation, and the World Health Organization. WHO https://apps.who.int/iris/handle/10665/39037
Henry NL, Hayes DF (2012) Cancer biomarkers. Mol Oncol 6:140–146
Article PubMed PubMed Central CAS Google Scholar
Simon R, Roychowdhury S (2013) Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov 12:358–369
Article PubMed CAS Google Scholar
Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D (2005) Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 353:1659–1672
Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, Roh JK (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360:1408–1417
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Coleman RL, Herzog TJ, Chan DW, Munroe DG, Pappas TC, Smith A, Zhang Z, Wolf J (2016) Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. Am J Obstet Gynecol 215:82–93
Article Google Scholar
U. S. Food & Drug Administration (2021) List of qualified biomarkers. F D A https://www.fda.gov/drugs/biomarker-qualification-program/list-qualified-biomarkers
James EK (2020) Carcinogenesis. An introduction to interdisciplinary toxicology. Handbook of toxicology pathology (second edition). Academic Press, p 97–110. https://doi.org/10.1016/B978-0-12-813602-7.00008-9
Handy DE, Castro R, Loscalzo J (2011) Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation 123:2145–2156
Article PubMed PubMed Central Google Scholar
Collins FS, Varmus H (2015) A new initiative on precision medicine. N Engl J Med 372:793–805
Nora MAA, Connor T, Ishwarlal J (2021) Genetics, epigenetic mechanism. StatPearls
World Health Organization (2022) Cancer fact sheet. WHO, Geneva. http://www.who.int/health-topics/cancer#tab=tab_1/ . Accessed in Feb 2023
Kirwan A, Utratna M, O’Dwyer ME, Joshi L, Kilcoyne M (2015) Glycosylation-based serum biomarkers for cancer diagnostics and prognostics. Biomed Res Int. https://doi.org/10.1155/2015/490531
Crandall BF, Lau HL (1981) Alpha-fetoprotein: a review. Crit Rev Clin Lab Sci 15:127–185
Mizejewski GJ (2018) Alpha-Fetoprotein (AFP) and gastric cancer: why is lethality more prevalent in AFP-secreting than non-secreting tumors? Cancer Ther Oncol Int J. https://doi.org/10.19080/CTOIJ.2018.09.555753
AlSalloom AAM (2016) An update of biochemical markers of hepatocellular carcinoma. Int J Health Sci 10:121–136
Google Scholar
Asahina Y, Tsuchiya K, Nishimura T, Muraoka M, Suzuki Y, Tamaki N, Yasui Y, Hosokawa T, Ueda K, Nakanishi H, Itakura J (2013) α-fetoprotein levels after interferon therapy and risk of hepatocarcinogenesis in chronic hepatitis C. Hepatol 58:1253–1262
Article CAS Google Scholar
Albrecht H, Carraway KL (2011) MUC1 and MUC4: switching the emphasis from large to small. Cancer Biother Radiopharm 26:261–271
PubMed PubMed Central CAS Google Scholar
Mukhopadhyay P, Chakraborty S, Ponnusamy MP, Lakshmanan I, Jain M, Batra SK (2011) Mucins in the pathogenesis of breast cancer: implications in diagnosis, prognosis and therapy. Biochim Biophys Acta 1815:224–240
Mudduwa LK, Wijayaratne GB, Peiris HH, Gunasekera SN, Abeysiriwardhana D, Liyanage N (2018) Elevated pre-surgical CA15-3: does it predict the short-term disease-free survival of breast cancer patients without distant metastasis? Int J Women Health 10:329–335
Duffy MJ, Evoy D, McDermott EW (2010) CA 15–3: uses and limitation as a biomarker for breast cancer. Clin Chim Acta 411:1869–1874
Perkins GL, Slater ED, Sanders GK, Prichard JG (2003) Serum tumor markers. Am Fam Physician 68:1075–1082
PubMed Google Scholar
Koprowski H, Steplewski Z, Mitchell K, Herlyn M, Herlyn D, Fuhrer P (1979) Colorectal carcinoma antigens detected by hybridoma antibodies. Somatic Cell Genet 5:957–971
Pall M, Iqbal J, Singh SK, Rana SV (2012) CA 19–9 as a serum marker in urothelial carcinoma. Urol Ann 4:98–101
Kim S, Park BK, Seo JH, Choi J, Choi JW, Lee CK, Chung JB, Park Y, Kim DW (2020) Carbohydrate antigen 19–9 elevation without evidence of malignant or pancreatobiliary diseases. Sci Rep 10:1–9
Ballehaninna UK, Chamberlain RS (2012) The clinical utility of serum CA 19–9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J Gastrointest Oncol 3:105–119
Kabel AM (2017) Tumor markers of breast cancer: new perspectives. J Oncol Sci 3:5–11
Vaidyanathan K, Vasudevan DM (2012) Organ specific tumor markers: what’s new? Indian J Clin Biochem 27:110–120
Nikhil TG, Dolly R, Staros EB (2021) CA 27–29: reference range, interpretation, collection and panels. Medscape https://emedicine.medscape.com/article/2087535-overview
Lin DC, Genzen JR (2018) Concordance analysis of paired cancer antigen (CA) 15–3 and 27.29 testing. Breast Cancer Res Treat 167:269–276
Muinao T, Boruah HP, Pal M (2018) Diagnostic and prognostic biomarkers in ovarian cancer and the potential roles of cancer stem cells - an updated review. Exp Cell Res 362:1–10
Menczer J, Ben-Shem E, Golan A, Levy T (2015) The significance of normal pre-treatment levels of CA125 (< 35 U/mL) in epithelial ovarian carcinoma. Rambam. Maimonides. Med J. https://doi.org/10.5041/RMMJ.10180
Pepin K, Carmen MD, Brown A, Dizon DS (2014) CA 125 and epithelial ovarian cancer: role in screening, diagnosis, and surveillance. Am J Hematol 10:22–29
Rustin GJ, Bast RC, Kelloff GJ, Barrett JC, Carter SK, Nisen PD, Sigman CC, Parkinson DR, Ruddon RW (2004) Use of CA-125 in clinical trial evaluation of new therapeutic drugs for ovarian cancer. Clin Cancer Res 10:3919–3926
Pignata S, Cannella L, Leopardo D, Bruni GS, Facchini G, Pisano C (2011) Follow-up with CA125 after primary therapy of advanced ovarian cancer: in favor of continuing to prescribe CA125 during follow-up. Ann Oncol 22:viii40–viii44
Bast RC Jr (2010) CA 125 and the detection of recurrent ovarian cancer: a reasonably accurate biomarker for a difficult disease. Cancer 116:2850–2853
Gold P, Freedman SO (1965) Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J Exp Med 121:439–462
Zhong W, Yu Z, Zhan J, Yu T, Lin Y, Xia ZS, Yuan YH, Chen QK (2015) Association of serum levels of CEA, CA199, CA125, CYFRA21-1 and CA72-4 and disease characteristics in colorectal cancer. Pathol Oncol Res 21:83–95
Saito G, Sadahiro S, Kamata H, Miyakita H, Okada K, Tanaka A, Suzuki T (2017) Monitoring of serum carcinoembryonic antigen levels after curative resection of colon cancer: cut off values determined according to preoperative levels enhance the diagnostic accuracy for recurrence. Oncology 92:276–282
Xie HL, Gong YZ, Kuang JA, Gao F, Tang SY, Gan JL (2019) The prognostic value of the postoperative serum CEA levels/preoperative serum CEA levels ratio in colorectal cancer patients with high preoperative serum CEA levels. Cancer Manag Res 11:7499–7511
Sørensen CG, Karlsson WK, Pommergaard HC, Burcharth J, Rosenberg J (2016) The diagnostic accuracy of carcinoembryonic antigen to detect colorectal cancer recurrence–a systematic review. Int J Surg 25:134–144
Wang Z, Wang W, Xu S, Wang S, Tu Y, Xiong Y, Mei J, Wang C (2016) The role of MAPK signaling pathway in the Her-2-positive meningiomas. Oncol Rep 36:685–695
Yan M, Schwaederle M, Arguello D, Millis SZ, Gatalica Z, Kurzrock R (2015) HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev 34:157–164
Furrer D, Paquet C, Jacob S, Diorio C (2018) The Human Epidermal Growth Factor Receptor 2 (HER2) as a prognostic and predictive biomarker: molecular insights into HER2 activation and diagnostic implications. Cancer Prognosis. https://doi.org/10.5772/intechopen.78271
Wang XY, Zheng ZX, Sun Y, Bai YH, Shi YF, Zhou LX, Yao YF, Wu AW, Cao DF (2019) Significance of HER2 protein expression and HER2 gene amplification in colorectal adenocarcinomas. World J Gastrointest Oncol 11:335–347
Ferraro S, Panteghini M (2019) Making new biomarkers a reality: the case of serum human epididymis protein 4. Clin Chem Lab Med 57:1284–1294
Lu R, Sun X, Xiao R, Zhou L, Gao X, Guo L (2012) Human epididymis protein 4 (HE4) plays a key role in ovarian cancer cell adhesion and motility. Biochem Biophys Res Commun 419:274–280
Zhu L, Zhuang H, Wang H, Tan M, Schwab CL, Deng L, Gao J, Hao Y, Li X, Gao S, Liu J (2016) Overexpression of HE4 (human epididymis protein 4) enhances proliferation, invasion and metastasis of ovarian cancer. Oncotarget 7:729–744
Moradi A, Srinivasan S, Clements J, Batra J (2019) Beyond the biomarker role: prostate-specific antigen (PSA) in the prostate cancer microenvironment. Cancer Metastasis Rev 38:333–346
Hong SK (2014) Kallikreins as biomarkers for prostate cancer. BioMed Res Int. https://doi.org/10.1155/2014/526341
Siemińska L, Borowski A, Marek B, Nowak M, Kajdaniuk D, Warakomski J, Kos-Kudła B (2018) Serum concentrations of adipokines in men with prostate cancer and benign prostate hyperplasia. Endokrynol Pol 69:120–127
Duffy MJ (2020) Biomarkers for prostate cancer: prostate-specific antigen and beyond. Clin Chem Lab Med 58:326–339
Tamhane S, Gharib H (2016) Thyroid nodule update on diagnosis and management. Clin Diabetes Endocrinol 2:1–10
Prpić M, Franceschi M, Romić M, Jukić T, Kusić Z (2018) Thyroglobulin as a tumor marker in differentiated thyroid cancer–clinical considerations. Acta Clin Croat 57:518–526
PubMed PubMed Central Google Scholar
Hasanbegovic L, Alicelebic S, Sljivo N (2015) Comparison of specific ovarian tumor markers by elecsys analyzer 2010. Acta Inform Med 23:86–89
Ueland FR, Desimone CP, Seamon LG, Miller RA, Goodrich S, Podzielinski I, Sokoll L, Smith A, Van Nagell Jr JR, Zhang Z (2011) Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstet Gynecol 117:1289–1297
Zhang Z, Chan DW (2010) The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev 19:2995–2999
Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, Gajewski W, Kurman R, Bast RC Jr, Skates SJ (2009) A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol 112:40–46
Anton C, Carvalho FM, Oliveira EI, Maciel GA, Baracat EC, Carvalho JP (2012) A comparison of CA125, HE4, risk ovarian malignancy algorithm (ROMA), and risk malignancy index (RMI) for the classification of ovarian masses. Clinics 67:437–441
Wang J, Gao J, Yao H, Wu Z, Wang M, Qi J (2014) Diagnostic accuracy of serum HE4, CA125 and ROMA in patients with ovarian cancer: a meta-analysis. Tumor Biol 35:6127–6138
Al Musalhi K, Al Kindi M, Al Aisary F, Ramadhan F, Al Rawahi T, Al Hatali K, Mula-Abed WA (2016) Evaluation of HE4, CA-125, risk of ovarian malignancy algorithm (ROMA) and risk of malignancy index (RMI) in the preoperative assessment of patients with adnexal mass. Oman Med J 31:336–344
Sharan RN (2009) 15 poly-ADP-ribosylation in cancer. In: Trygve Tollefsbol (ed) Cancer Epigenetics. CRC Press, Florida, pp 265–279
Chanu TM (2022) PhD thesis entitled Clinical correlation between cancer regression and selected genetic and epigenetic biomarkers in patients undergoing cancer therapy. NEHU, Shillong, India
Sharan RN, Devi BJ, Humtsoe JO, Saikia JR, Kma L (2005) Detection and quantification of poly-ADP-ribosylated cellular proteins of spleen and liver tissues of mice in vivo by slot and Western blot immunoprobing using polyclonal antibody against mouse ADP-ribose polymer. Mol Cell Biochem 278:213–221
Devi BJ, Schneeweiss FHA, Sharan RN (2005) Negative correlation between poly-ADP-ribosylation of spleen cell histone proteins and initial duration of dimethylnitrosamine exposure to mice in vivo measured by Western blot immunoprobe assay: a possible biomarker for cancer detection. Cancer, Detect Prev 29:66–71
Saikia JR, Schneeweiss FHA, Sharan RN (1998) Effects of chronic low-dose arecoline administration on the macromolecular components of bone marrow and spleen cells of mice. Cancer J 11:94–98
CAS Google Scholar
Kma L, Sharan RN (2014) Dimethylnitrosamine induced reduction in the level of poly- ADP ribosylation of histone proteins of blood lymphocytes – a sensitive and reliable biomarker for early detection of cancer. Asia Pac J Cancer Prev 15:6429–6436
Lakadong RO, Kataki AC, Sharan RN (2010) ADP-ribose polymer - a novel and general biomarker of human cancers of head & neck, breast, and cervix. Mol Cancer. https://doi.org/10.1186/1476-4598-9-286
Hoxhaj G, Manning BD (2020) The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer 20:74–88
Degan SE, Gelman IH (2021) Emerging roles for AKT isoform preference in cancer progression pathways. Mol Cancer Res 19:1251–1257
Turner KM, Sun Y, Ji P, Granberg KJ, Bernard BL, Cogdell DE, Zhou X, Yli-Harja O, Nykter M, Shmulevich I (2015) Genomically amplified Akt3 activates DNA repair pathway and promotes glioma progression. Proc Natl Acad Sci 112:3421–3426
Gallyas F, Sumegi B, Szabo C (2020) Role of Akt activation in PARP inhibitor resistance in cancer. Cancers 12:532–540
Guo K, Tang W, Zhuo H, Zhao G (2019) Recent advance of Akt inhibitors in clinical trials. ChemistrySelect 4:9040–9044
Santana dos Santos E, Lallemand F, Burke L, Stoppa-Lyonnet D, Brown M, Caputo SM, Rouleau E (2018) Non-coding variants in BRCA1 and BRCA2 genes: potential impact on breast and ovarian cancer predisposition. Cancers. https://doi.org/10.3390/cancers10110453
Huang F, Goyal N, Sullivan K, Hanamshet K, Patel M, Mazina OM, Wang CX, An WF, Spoonamore J, Metkar S, Emmitte KA (2016) Targeting BRCA1-and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acid Res 44:4189–4199
Jin TY, Park KS, Nam SE, Yoo YB, Park WS, Yun IJ (2022) BRCA1/2 serves as a biomarker for poor prognosis in breast carcinoma. Intl J Mol Sci 23:3754–3768
Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC (2000) Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J 19:1148–1156
Dutto I, Scalera C, Prosperi E (2018) CREBBP and p300 lysine acetyl transferases in the DNA damage response. Cell Mol Life Sci 75:1325–1338
Karimian A, Ahmadi Y, Yousefi B (2016) Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 42:63–71
Bertoli C, Skotheim JM, De Bruin RA (2013) Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 14:518–528
Abbas T, Dutta A (2009) p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9:400–414
Moor NA, Lavrik OI (2018) Protein–protein interactions in DNA base excision repair. Biochemistry 83:411–422
PubMed CAS Google Scholar
Weiss RH, Borowsky AD, Seligson D, Lin PY, Dillard-Telm L, Belldegrun AS, Figlin RA, Pantuck AD (2007) p21 is a prognostic marker for renal cell carcinoma: implications for novel therapeutic approaches. J Urol 177:63–69
Nongrum S, Vaiphei ST, Keppen J, Ksoo M, Kashyap E, Sharan RN (2017) Identification and preliminary validation of radiation response proteins(s) in human blood for a high throughput molecular biodosimetry technology for the future. Genome Integr. https://doi.org/10.4103/2041-9414.198910
Das JK, Xiong X, Ren X, Yang JM, Song J (2019) H eat shock proteins in cancer immunotherapy J. Oncol. https://doi.org/10.1155/2019/3267207
Ciocca DR, Calderwood SK (2005) Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10:86–103
Lianos GD, Alexiou GA, Mangano A, Rausei S, Boni L, Dionigi G, Roukos DH (2015) The role of heat shock proteins in cancer. Cancer Lett 360:114–118
Moon A, Bacchini P, Bertoni F, Olvi LG, Santini-Arawo E, Kim YW, Par YK (2010) Expression of heat shock proteins in osteosarcomas. Pathology 42:421–425
Selleck MJ, Senthil M, Wall NR (2017) Making meaningful clinical use of biomarkers. Biomark Insights. https://doi.org/10.1177/1177271917715236
Wang W, Xu X, Tian B, Wang Y, Du L, Sun T, Shi Y, Zhao X, Jing J (2017) The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta 470:51–55
Schüler-Toprak S, Treeck O, Ortmann O (2017) Human chorionic gonadotropin and breast cancer. Int J Mol Sci. https://doi.org/10.3390/ijms18071587
Tian T, Li X, Zhang J (2019) mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int J Mol Sci 20:755–764
Conciatori F, Ciuffreda L, Bazzichetto C, Falcone I, Pilotto S, Bria E, Cognetti F, Milella M (2018) mTOR cross-talk in cancer and potential for combination therapy. Cancers 10:23–34
Zou Z, Tao T, Li H, Zhu X (2020) mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci 10:1–10
Lee HK, Lee DS, Park JC (2015) Nuclear factor IC regulates E-cadherin via control of KLF4 in breast cancer. BMC Cancer 15:1–11
Grabowska MM, Elliott AD, DeGraff DJ, Anderson PD, Anumanthan G, Yamashita H, Sun Q, Friedman DB, Hachey DL, Yu X, Sheehan JH (2014) NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression. Mol Endocrinol 28:949–964
Fane M, Harris L, Smith AG, Piper M (2017) Nuclear factor one transcription factors as epigenetic regulators in cancer. Int J Cancer 140:2634–2641
Yang C, Chng KR (2012) Abstract C2: Nuclear factor I/C collaborates with androgen receptor in transcription regulation in prostate cancer. Cancer Res. https://doi.org/10.1158/1538-7445.PRCA2012-C2
Zenker M, Bunt J, Schanze I, Schanze D, Piper M, Priolo M, Gerkes EH, Gronostajski RM, Richards LJ, Vogt J, Wessels MW (2019) Variants in nuclear factor I genes influence growth and development. Am J Med Gen 181:611–626
Carmeliet P (2005) VEGF as a key mediator of angiogenesis in cancer. Oncology 69:4–10
Hegde PS, Wallin JJ, Mancao C (2018) Predictive markers of anti-VEGF and emerging role of angiogenesis inhibitors as immunotherapeutics. Semin Cancer Biol 52:117–124
Kulapaditharom B, Boonkitticharoen V, Sritara C (2012) Plasma vascular endothelial growth factor dysregulation in defining aggressiveness of head and neck squamous cell carcinoma. J Oncol 15:12–20
Garcia J, Hurwitz HI, Sandler AB, Miles D, Coleman RL, Deurloo R, Chinot OL (2020) Bevacizumab (Avastin®) in cancer treatment: a review of 15 years of clinical experience and future outlook. Cancer Treat Rev 86:102–117
Download references
This research was financially supported by grants from (a) International Atomic Energy Agency, Vienna under research contract # 22218 to RNS and (b) DRS-III scheme of the UGC to the Department of Biochemistry. TMC gratefully acknowledges junior and senior research fellowship grants to support her doctoral study under the “Innovation in Science Pursuit for Inspired Research (INSPIRE)” Fellowship scheme of the Government of India.
Author information
Authors and affiliations.
Radiation & Molecular Biology Unit, Department of Biochemistry, North-Eastern Hill University, Shillong, 793022, India
Tongbram Malemnganbi Chanu & R. N. Sharan
Cancer & Radiation Countermeasures Unit, Depatment of Biochemistry, North-Eastern Hill University, Shillong, 793022, India
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to R. N. Sharan .
Ethics declarations
Conflict of interest.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Chanu, T.M., Kma, L. & Sharan, R.N. Cancer Biomarkers: Status and Its Future Direction. Indian J Surg 85 , 1323–1335 (2023). https://doi.org/10.1007/s12262-023-03723-1
Download citation
Received : 19 January 2023
Accepted : 13 February 2023
Published : 20 February 2023
Issue Date : December 2023
DOI : https://doi.org/10.1007/s12262-023-03723-1
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Cancer biomarker
- Personalized cancer therapy
- Find a journal
- Publish with us
- Track your research
REVIEW article
Biomarkers in colorectal cancer: the role of translational proteomics research.

- 1 Medical Sciences Postgraduate Program, School of Medicine, University of Brasilia, Brasília, Brazil
- 2 UniCeub—Centro Universitário Do Distrito Federal, Translational Medicine Group, School of Medicine, Brasilia, Brazil
- 3 Department of Cell Biology, Institute of Biology, University of Brasilia, Brasilia, Brazil
- 4 Metabolomics and Bioanalysis Center, San Pablo CEU University, Madrid, Spain
Colorectal cancer is one of the most common cancers in the world, and it is one of the leading causes of cancer-related death. Despite recent progress in the development of screening programs and in the management of patients with colorectal cancer, there are still many gaps to fill, ranging from the prevention and early diagnosis to the determination of prognosis factors and treatment of metastatic disease, to establish a personalized approach. The genetic profile approach has been increasingly used in the decision-making process, especially in the choice of targeted therapies and in the prediction of drug response, but there are still few validated biomarkers of colorectal cancer for clinical practice. The discovery of non-invasive, sensitive, and specific biomarkers is an urgent need, and translational proteomics play a key role in this process, as they enable better comprehension of colorectal carcinogenesis, identification of potential markers, and subsequent validation. This review provides an overview of recent advances in the search for colorectal cancer biomarkers through proteomics studies according to biomarker function and clinical application.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer among adults and is the third leading cause of cancer-related death in the United States ( 1 ). Most colorectal cancers occur sporadically and are characterized by a sequenced carcinogenesis process that involves the progressive accumulation of mutations in a period that lasts on average 10–15 years ( 2 – 5 ). This long evolution interval allows for the successful application of screening, early detection of cancer, and removal of premalignant lesions (adenomas), leading to a reduction in incidence and mortality ( 5 – 8 ). Despite the opportunity for early diagnosis, ~20–25% of CRC cases are diagnosed at stage IV, when the patients have already presented with distant metastasis and the 5-year survival rate is <10%. In contrast, the 5-year survival for patients with early localized disease, when surgical resection is possible, may be as high as 90% ( 9 , 10 ).
The current gold standard screening strategy is through a colonoscopy. The guidelines recommend that individuals aged 45 years and older with an average risk of CRC undergo regular screening ( 8 ). However, colonoscopies have poor patient compliance. The procedure is expensive and invasive and carries risks, such as hemorrhage, colonic perforation, and cardiorespiratory complications. Other reasons for low adherence are related to a preoccupation with pudency, procedure discomfort, and bowel preparation ( 11 ). The most frequently used non-invasive screening method is the guaiac fecal occult blood test (gFOBT), based on the identification of hemoglobin peroxidase activity in the stool. Although FOBT is an easy and cost-effective method for screening CRC, it has relatively poor selectivity and sensitivity, resulting in high rates of both false positives and false negatives ( 4 , 5 ).
Therefore, alternative cost-effective, non-invasive, easily measurable, and accurate screening procedures are urgently required for CRC screening. Thus, the clinical applications of biomarkers in CRC are not only needed for the early detection of the disease but are also essential for prognostic stratification, surveillance, and therapy selection ( Figure 1 ) ( 12 – 14 ). The increasing emergence of adjuvant and neoadjuvant therapy approaches results in an urgent need for predictive biomarkers that guide the decision-making process ( 12 ). An example of the importance of predictive biomarkers is how treatment with drugs can antagonize the epidermal growth factor receptor (EGFR) in patients with KRAS-wild-type tumors. The discovery of this targeting therapy made the determination of KRAS status a mandatory step for the adequacy of chemotherapy in patients with advanced colorectal cancer ( 15 ).
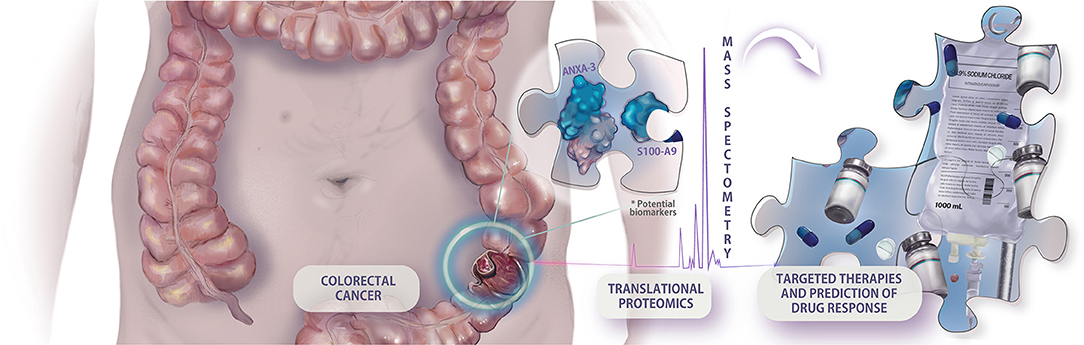
Figure 1 . Example of hypothetical application of translational proteomic research in colorectal cancer approach. The prospection of new predictive biomarkers is cardinal to the implementation of an integrative and personalized medicine, making possible the individual assessment of targeted therapies, and drug response.
Recent progress in genomics, transcriptomics, proteomics, and metabolomics has expanded the number of candidate biomarkers and led to better comprehension of the progression of colorectal cancer as well as the identification of molecular signatures ( 16 – 22 ).
Dysplastic and neoplastic tissues regulate the expression of proteins and generate protein profiles that may be associated with the progression of these lesions in many different and interacting signaling pathways ( 23 ). Proteomics represents a large number of approaches employed for large-scale recognition, measurement, characterization, and analysis of proteins. The majority of studies on biomarker discovery employ quantitative mass spectrometry-based techniques for the identification and validation of dysregulated proteins as disease biomarker candidates ( 24 ). Translational proteomics research emphasizes the translation of general proteomics science to determine protein expression profiles that generate pathogenic phenotype variations and contribute to clinical practice ( 15 ).
This review aims to provide an overview of recent advances in mass spectrometry-based proteomics in the search for protein biomarkers of CRC with the potential for clinical application according to biomarker functions: diagnostic, predictive, or prognostic.
Diagnostic Biomarkers
A diagnostic biomarker can be defined as a biological characteristic that detects or suggests the presence of a disease or condition of interest or identifies an individual with a subtype of the disease ( 25 ).
It is well-established that colorectal cancer screening strategies that lead to the identification and removal of adenomatous polyps and other premalignant lesions result in a decrease in CRC mortality ( 26 ). Colonoscopies are the only screening method that can identify and remove precancerous polyps; however, the exam requires bowel preparation and dietary modification, it is operator dependent, and it has been associated with major complications, such as cardiopulmonary events, gastrointestinal bleeding, and perforation ( 27 ). Perforation is the most frequent major complication, occurring in 0.016–0.8% of diagnostic examinations and up to 5% of therapeutic colonoscopies ( 28 ).
Some studies report the non-attendance rate of colonoscopies to be 10–20% after a positive fecal occult blood test ( 29 , 30 ). The main factors associated with non-adherence with colonoscopies are laxative bowel preparation, lack of awareness of the significance of screening, and concerns about embarrassment, modesty, and dignity ( 31 ). Plumb et al. ( 32 ) evaluated the explanations for colonoscopy non-participation, and ~30% of the patients addressed the unwillingness to undergo the test as the major barrier to go through with the whole screening program.
Non-invasive methods such as fecal immunochemical tests, gFOBT, and stool DNA tests can be used for regular screening, but positive results should be followed up with timely colonoscopy ( 8 ). The current fecal occult blood test methods are more easily accepted by participants in population screening programs; however, they are subject to various interfering factors with some causes of false-negative, false-positive results, and low sensitivity rates for detecting colon polyps ( 33 – 35 ) Therefore, early, non-invasive, specific, and sensitive biomarkers are still required for screening strategies in colorectal cancer.
Many proteomic approaches have been used in the search for potential diagnostic biomarkers. Ghazanfar et al. ( 36 ) performed two-dimensional gel electrophoresis coupled with mass spectrometry for the expression profiling of proteins extracted from freshly frozen human colorectal cancer tissue specimens (12 patients) and neighboring non-tumor tissue, and they demonstrated the upregulation of some proteins, such as actin beta-like 2 (ACTBL2), in colorectal cancer. Hao et al. ( 37 ) used high-resolution Fourier transform mass spectrometry to evaluate 22 pairs of cancerous and adjacent normal tissue specimens that were gathered from 22 individuals and revealed an overexpression of dipeptidase 1 (DPEP1) in colorectal tumor tissue.
Formalin-fixed paraffin-embedded (FFPE) tissues can also be used in proteomics approaches, allowing access to archival samples, allowing usage of larger cohorts and more robust analyses, and optimizing the follow-up data of patients' clinical conditions. Quesada-Calvo et al. ( 23 ) analyzed 76 formalin-fixed paraffin-embedded colorectal tissues from early CRC stages (pT1N0M0 and pT2N0M0), as well as normal or inflamed mucosa, by label-free proteomics, and different expression levels of olfactomedin-4 (OLFM4), kininogen-1 (KNG1), and transport protein Sec24C (Sec24C) were observed in the early CRC stages compared to normal and premalignant tissues. Although the experiment was performed with liquid chromatography-mass tandem mass spectrometry (LC-MS/MS), the results were also validated by immunohistochemistry of these annotated effectors. Yamamoto et al. ( 38 ) also used formalin-fixed and paraffin-embedded (FFPE) CRC tissue to perform liquid chromatography (LC)/mass spectrometry (MS) based on a global proteomic approach, revealing higher expression levels of cyclophilin A, annexin A2, and aldolase A in cancer compared to non-cancer regions ( 38 ).
Blood-based biomarkers are potentially the best matrices for early diagnosis and surveillance of colorectal cancer because the specimens can be obtained easily by a non-invasive method with minimal cost and risk ( 24 , 39 ). Ivancic et al. ( 40 ) used targeted liquid chromatography-tandem mass spectrometry to analyze blood from 213 healthy individuals and 50 patients with non-metastatic CRC. This approach resulted in a panel of five proteins (leucine-rich alpha-2-glycoprotein 1, EGFR, inter-alpha-trypsin inhibitor heavy-chain family member 4, hemopexin, and superoxide dismutase 3) with good performance for CRC detection, which present 89% specificity at over 70% sensitivity in the validation set. Bhardwaj et al. ( 41 ) also proposed a protein panel for the early detection of CRC, utilizing an approach with liquid chromatography/multiple reaction monitoring-mass spectrometry and a subsequent proximity extension assay to analyze plasma from 96 CRC patients and 94 controls. They demonstrated promising CRC-screening performance of a five-marker blood-based profile consisting of mannan binding lectin serine protease 1, osteopontin, serum paraoxonase lactonase 3, transferrin receptor protein 1, and amphiregulin.
Yu et al. ( 42 ) used magnetic beads and matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry to analyze 127 CRC serum samples and 90 healthy control serum samples. The protein serine/threonine kinase 4 (STK4 or MST1) was identified by tandem mass spectrometry (MS/MS) and validated with Western blotting and an enzyme-linked immunosorbent assay (ELISA). They demonstrated a downregulation of MST1 in CRC patients, with a sensitivity of 92.3% and specificity of 100% in the diagnosis of colorectal cancer when gathered with carcinoembryonic antigen and FOBT. Their work also implied that MST1 could be a predictive marker for distant metastasis ( 42 ).
Fan et al. ( 43 ) also conducted a study with serum samples that were analyzed by a combination of high-performance liquid chromatography and mass spectrometry and further validation with Western blotting. They verified an upregulation of macrophage mannose receptor 1 (MRC1) and S100 calcium-binding protein A9 (S100A9) in colorectal cancer. Members of the serpin family, such as SERPINA1 (alpha-1-antitrypsin, A1AT), SERPINA3 (alpha-1-antichymotrypsin, AACT), and SERPINC1 (antithrombin-3, AT-III), have also been described as potential biomarkers of adenomatous polyps and colorectal carcinomas through analyses of serum samples by multiplexed quantification with an isobaric tag for relative and absolute quantitation (iTRAQ) ( 44 ).
Despite the expansion of MS-based proteomics research and the large number of diagnostic biomarker candidates ( Table 1 shows some examples of candidates for diagnostic protein biomarkers), none of them were successfully translated into clinical practice. This probably occurs due to the difficulty of validating the possible biomarkers in large cohorts and comparing the results with the current screening methods. However, the continuation of proteomic research is essential because, certainly, there is a space in the CRC screening that needs to be filled by reliable biomarkers.
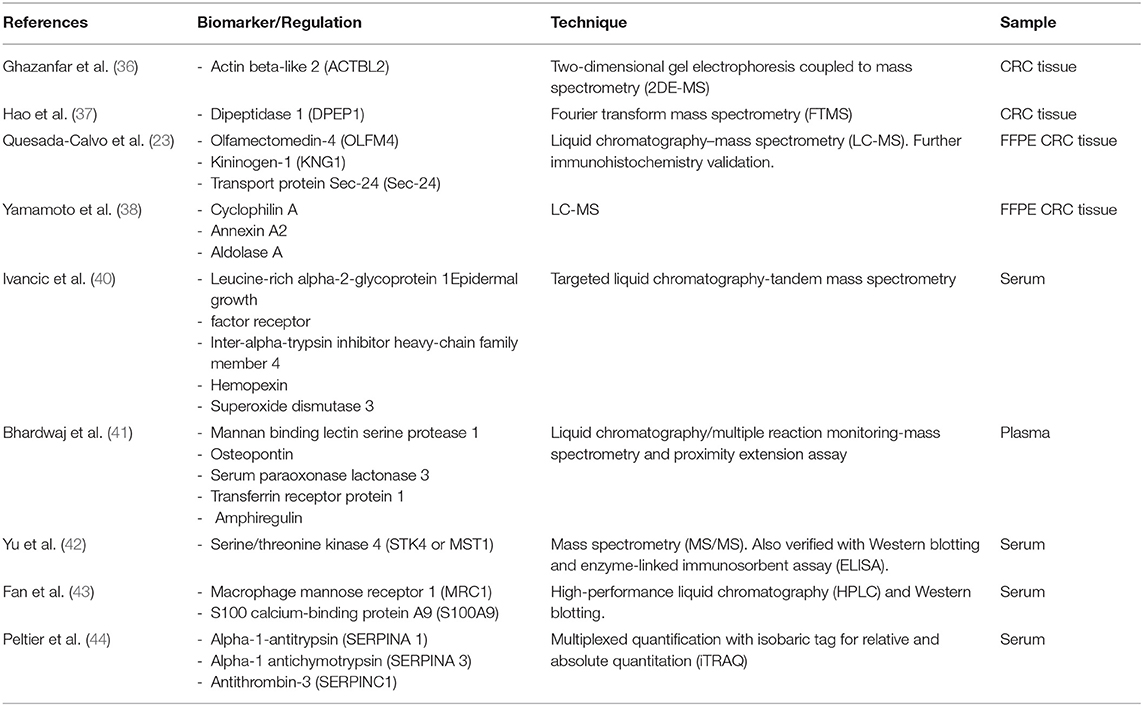
Table 1 . Examples of candidate diagnostic biomarkers.
Predictive Biomarkers
The predictive biomarkers are used to indicate the response to a specific treatment and to guide the decision-making process. The prospection of new predictive biomarkers is crucial to the evolution of the management of patients with colorectal cancer in the near future, and proteomics represent a powerful strategy for the discovery and implementation of personalized approaches. The increasing number of chemotherapy and immunotherapy drugs and the emergence of target therapies make it necessary to discover some response parameters and monitoring evaluations ( 45 , 46 ).
Concerning the individualized and integrative treatment of patients with colorectal cancer, the understanding of the mechanism underlying chemotherapy resistance is a prerequisite to overcome the resistance and improve the efficacy of chemotherapy. In addition, the identification of good-responder patients is also important to guide and improve personalized therapies. Wang et al. ( 47 ) correlated the capacity of proteomic, genomic, and transcriptomic profiles to predict drug sensitivity. Forty-four CRC cell lines were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based shotgun proteomics and compared against 90 colorectal cancer primary tumor specimens and 60 normal tissue biopsies. The proteomic profile was compared on mutations, DNA copy number, and mRNA expression, and the results showed that proteomic data tended to exhibit better potential for predicting sensitivity to 5-fluorouracil, SN-38, erlotinib, regorafenib, and oxaliplatin when compared to genomic and transcriptomic profiles ( 47 ).
Guo et al. ( 48 ) investigated protein elements that might be implicated in oxaliplatin resistance by comparing the proteome between oxaliplatin-sensitive HT-29 wild-type cells and oxaliplatin-resistant HT-29 cells using 2D gel electrophoresis followed by MALDI TOF/TOF mass spectrometry. It was observed that poly(C)-binding protein 1 (PCBP1) expression was significantly more elevated in tumor samples from oxaliplatin-refractory patients than in those from responsive patients, suggesting that PCBP1 is a protein marker of oxaliplatin resistance in colorectal cancer cell cultures.
Martin et al. ( 49 ) evaluated the response to vascular endothelial growth factor inhibitor (bevacizumab) in patients with metastatic colorectal cancer through the analysis of pretreatment serum from 23 patients. 2D difference gel electrophoresis (2D-DIGE) was performed, followed by LC-MS/MS, which identified 68 differentially expressed proteins between responders and non-responders. Three proteins, apolipoprotein E (APOE), angiotensinogen (AGT), and vitamin D-binding protein (DBP), were chosen for validation through immunohistochemistry and enzyme-linked immunosorbent assay (ELISA) and were correlated with better survival outcomes in patients treated with chemotherapy and bevacizumab ( 49 ).
The response to EGFR-targeted therapies was also evaluated by Katsila et al. ( 50 ) through a quantitative proteomic analysis of the plasma of patients with metastatic colorectal cancer compared with the 3D colorectal cancer spheroid secretome (isogenic cells SW48) of patients treated with cetuximab. They showed that the plasma level of phosphorylated-EGFR (pEGFR) was associated with sensitivity to cetuximab therapy, suggesting that pEGFR could be a predictive drug-response biomarker ( 50 ).
An expanding research area due to tailored-made therapy for patients with colorectal cancer is the therapeutic targets in anti-tumor immunity ( 51 ). Studies with immune checkpoint-inhibiting drugs, such as those directed against cytotoxic T-lymphocyte antigen 4 (CTLA-4) and programmed death-1 receptor (PD1) and its ligand PD-L1, have demonstrated promising results in the therapy of patients with metastatic colorectal cancer ( 52 – 54 ).
Until now, the best indicator of responsiveness to immunotherapy in patients with colorectal cancer seems to be mismatch repair deficiency ( 55 ). Repair system deficiency leads to a high burden of somatic mutations, which increases the immunogenicity ( 51 ).
Furthermore, tumors with high microsatellite instability (MSI-H) present a dense Th1 lymphocytic infiltration and a cytokine-rich microenvironment that is related to the highly upregulated expression of multiple immune checkpoint proteins ( 56 ). Unfortunately, the patients with MSI-H tumors represent only a subgroup of the patients with colorectal cancer, and the likelihood of mismatch repair deficiency varies according to the stage of the disease, reaching 4–5% in the metastatic disease. In addition, not all patients with MSI-H tumors respond to immunotherapy ( 57 ).
Therefore, a complete understanding of the response of the immune system to MSI-H is crucial to optimizing the immunotherapy approach. Some studies have demonstrated promising prognostic biomarkers, such as the expression of heat shock protein 110 and protein ß2-microglobulin, to stratify patients with MSI-H CRC according to prognosis ( 58 , 59 ).
In this scenario, the application of mass spectrometry-based immune-proteomic methods is a powerful tool in the search for overexpressed immunogenic proteins that could be new targets of immunotherapeutic development. Yang et al. ( 60 ) used mass spectrometry to evaluate antibody-reactive proteins, and this was followed by Western blotting and immunohistochemistry validation. Their experiment described differential expression of proteasome subunit alpha type 1 (PSA1), leucine aminopeptidase 3 (LAP3), annexin A3 (ANXA3), and maspin (serpin B5), demonstrating a proteomic profile of antibody-inducing cancer-associated immunogens ( 60 ). In another study with an immuno-proteomic approach by the same group, overexpression of olfactomedin 4, CD11b, and integrin alpha-2 was identified in the tumor tissue of patients with colorectal cancer with liver metastases ( 61 ).
The current treatment of locally advanced rectal cancer (stages II and III) is neoadjuvant chemoradiation followed by surgery ( 62 ). The main role of neoadjuvant therapy is local tumor control, but, in ~10–20% of patients, a pathologic complete response is observed. This fact allows for the possibility of a selective surgical approach, which was described in Habr-Gama et al. ( 63 ). One of the most challenging issues in the modern management of patients with rectal cancer is to predict the response to neoadjuvant therapy. Recently, Chauvin et al. ( 64 ) highlighted different protein signatures in patients who underwent neoadjuvant therapy in a study on mass spectrometry of formalin-fixed paraffin-embedded tumor biopsies. The researchers identified that interferon-induced protein with tetratricopeptide repeats 1 (IFIT1), FAST kinase domains 2 (FASTKD2), phosphatidylinositol-5-phosphate 4-kinase type-2 beta (PIP4K2B), AT-rich interactive domain-containing protein 1B (ARID1B), and solute carrier family 25 member 33 (SLC25A33) were overexpressed in the tumor tissue of the initial biopsy from patients who achieved complete response to neoadjuvant chemoradiotherapy. In the non-responder group, they identified that caldesmon 1 (CALD1), carboxypeptidase A3 (CPA3), beta-1,3-galactosyltransferase 5 (B3GALT5), CD177, and receptor-interacting serine/threonine-protein kinase 1 (RIPK1) were overexpressed ( 64 ).
The predictive protein biomarkers face the same problem of slow translation to clinical application as the diagnostic biomarkers. Table 2 shows some examples of candidates for predictive protein biomarkers. The pursuit for new biomarkers maintains a central role in the development of the integrative management of CRC patients because it is crucial to determine the responses to neoadjuvant and adjuvant therapies. Mass spectrometry's ability to detect low-abundance elements makes this technique a powerful tool for prospecting these potential biomarkers.
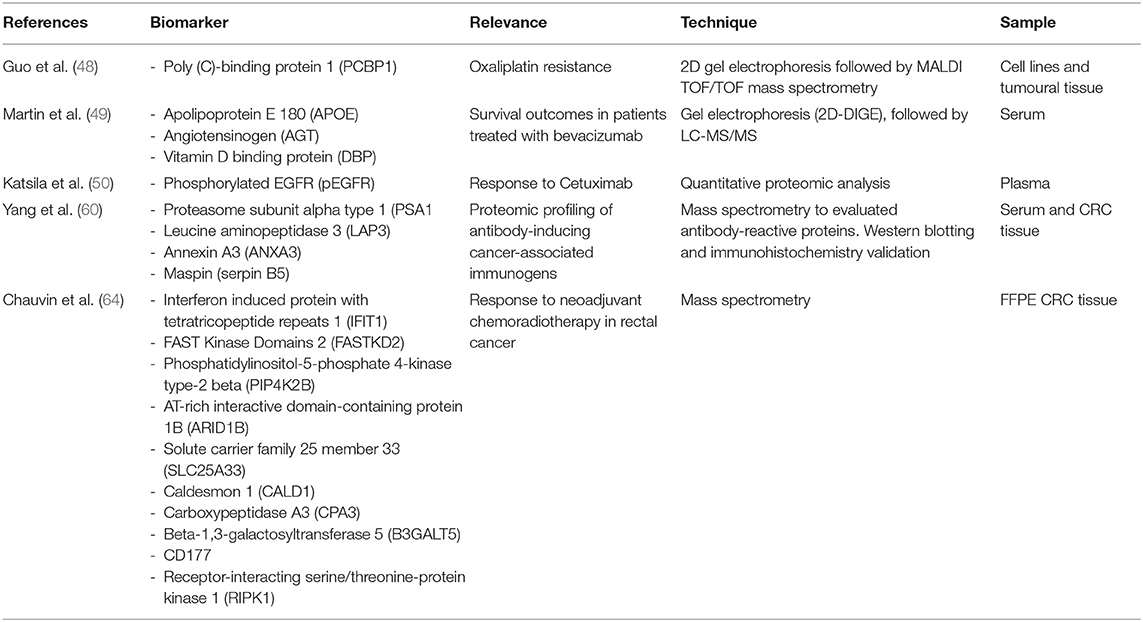
Table 2 . Examples of candidate predictive biomarkers.
Prognostic Biomarkers
A prognostic biomarker can be defined as a biological characteristic that gives information about the patient's overall cancer outcome, independent of therapy ( 65 ). The current staging strategy for colorectal cancer is the TNM system, which consists of the analysis of tumor depth of invasion (T), nodal involvement (N), and presence of metastasis (M) ( 66 ). The overall prognosis is determined by a combination of clinical and pathologic variables; however, the prognosis can be different between patients in the same stage, and, in some cases, patients at early stages can present poorer outcomes than patients at advanced stages. These variations are the result of a complex process of colorectal carcinoma (CRC) pathogenesis that involves multistep molecular pathways, initiated by genetic and epigenetic events ( 19 ).
The main prognostic biomarker used in clinical practice is carcinoembryonic antigen (CEA), a high-molecular-weight glycoprotein expressed in embryonic tissue and colorectal malignancies. This antigen was discovered in 1965, but it remains the most widely used blood-based biomarker for CRC. Elevated levels are associated with cancer progression and can indicate recurrence after surgery. However, high CEA levels are not specific to CRC and can also be found in other malignancies and inflammatory conditions, such as inflammatory bowel disease, liver disease, and pancreatitis ( 67 , 68 ).
Recently, other parameters have been used to determine the prognosis. The effect of microsatellite instability (MSI) and BRAF mutation on survival in colorectal carcinoma was elucidated, and these genetic markers already have clinical applications ( 19 ). Despite these recent advances, additional prognostic biomarkers are urgently needed to optimize the management and follow-up of colorectal cancer patients.
The presence of metastases represents the main unfavorable prognostic factor in patients with colorectal cancer. The estimated 5-year survival for stage IV patients is ~8% ( 69 ). The major site of metastases in colorectal cancer is the liver, occurring in 20–35% of patients at the time of diagnosis and in nearly 70% of patients during the course of the disease ( 70 ). Marfà et al. ( 71 ) used a high-throughput proteomic technique to predict 5-year survival in patients with colorectal cancer who developed liver metastases. Human hepatic tumor samples were analyzed by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI), and a classification and regression tree analysis was done posteriorly. This approach allowed the identification of four relevant protein peaks and the construction of an algorithm that revealed an excellent diagnostic accuracy in differentiating mild from severe colorectal liver metastases patients ( 71 ).
Recently, Kirana et al. ( 72 ) performed a combination of laser microdissection, 2D-DIGE and MALDI-TOF MS to identify proteins associated with colorectal cancer spread. Initially, laser microdissection was applied to isolate cancer cells from primary colorectal tumors of stage II patients in two distinct groups: (i) patients who presented metastases within 5 years of initial surgical intervention and (ii) patients who did not present metastases within 5 years of initial surgical intervention. Then, 2D-DIGE (a technique that uses fluorescent dyes to label different conditions) and MALDI-TOF were used to identify the global profile of proteins, with posterior validation achieved through tissue microarray (TMA) immunohistochemistry. The expression of HLAB, 14-3-3β protein, a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS2), latent transforming growth factor beta binding protein 3 (LTBP3), nucleoside diphosphate kinase 2 (NME2), and jagged 2 protein (JAG2) was associated with clinical pathological parameters related to tumor progression, invasion, and metastasis ( 72 ).
Zhu et al. ( 73 ) used another approach, based on magnetic bead-based fractionation coupled with mass spectrometry, to compare serum samples from patients with metachronous liver metastases vs. patients without recurrence or metastases for at least 3 years after radical colorectal surgery. Serum proteomic fingerprinting was done, and it exhibited a promising value for predicting metachronous liver metastases in patients who underwent radical resection of colorectal cancer. The peptides were recognized as fragments of alpha-fetoprotein, complement C4-A, fibrinogen alpha, eukaryotic peptide chain release factor GTP-binding subunit ERF3B, and angiotensinogen ( 73 ).
The collagen proteins seem to be promising candidates as biomarkers in the metastatic scenario of colorectal cancer. A recent MS-based proteomic approach compared colorectal liver metastasis tissues with healthy adjacent liver tissues, demonstrating the upregulation of 19 of 22 collagen-α chains in colorectal liver metastasis tissue. Posterior validation with immunohistochemistry showed significant upregulation of collagen type XII in the metastatic context ( 74 ).
Some studies have also demonstrated the possibility of detecting colorectal liver metastases through the identification of collagen peptides in urine ( 75 , 76 ). Urine is an interesting potential source of biomarkers as this biological fluid is easy to obtain non-invasively ( 24 ). An example of a promising application of this method is the measurement of the urinary prostaglandin metabolite PGE-M. PGE-M is the major urinary metabolite of prostaglandin E2, which plays an important role in mediating the effects of cyclooxygenase-2 in colorectal carcinogenesis. Elevated urinary levels of PGE-M seem to be correlated with advanced adenomas and an elevated risk of colorectal cancer ( 77 – 81 ).
The determination of nodal status in CRC is another point that requires new candidate prognostic biomarkers. Lymph node involvement results in poor prognosis, reducing the 5-year survival rate from 70 to 80% in patients with node-negative disease to 30%−60% in those with node-positive disease ( 82 ). The current non-invasive imaging methods used for the preoperative detection of lymph node metastasis, such as computed tomography, magnetic resonance imaging, and endorectal ultrasound, have low accuracy rates ( 83 ).
Non-invasive methods to predict the nodal status could improve the management of patients with colorectal cancer, guiding the indication of chemotherapy or the extension of the surgery. Mori et al. ( 84 ), in a recent study, used isobaric tags for relative and absolute quantitation (iTRAQ) as part of a proteomic analysis that identified 60 differentially expressed proteins specifically related to lymph node metastasis in patients with colorectal cancer. The validation process by immunohistochemistry revealed that heat shock protein 47 (HSP47) expression in colorectal cancer tissue was significantly higher than in adjacent normal colonic mucosa ( 84 ).
In another study by the same group, iTRAQ was used in a comparative proteomics approach, demonstrating that a high level of ezrin protein was an independent predictor of lymph node metastasis in colorectal cancer ( 85 ). The ezrin protein seems to occupy an important place in the carcinogenesis process, being described as a biomarker candidate for the progression and prognosis of gastrointestinal cancers and a target for anti-metastatic therapy ( 86 , 87 ). Furthermore, some studies associate the upregulation of ezrin expression with rectal cancer recurrence and tumor aggressiveness ( 88 , 89 ).
One of the most concerning points in the postoperative follow-up of CRC is recurrence detection. After surgery with curative intent, 30–40% of the patients present locoregional recurrence or distant metastasis ( 90 ). Regarding the discovery of prognostic biomarkers of colorectal cancer recurrence, Clarke et al. ( 91 ), in a recent study, used a reverse phase protein array to unveil the functional proteome in 263 colorectal cancer tumor samples from patients treated at MD Anderson Cancer Center and 462 primary tumor tissue from The Cancer Genome Atlas archived colorectal tumor bank. On multivariate analysis, eight proteins demonstrated significant prognostic factors for tumor recurrence: collagen VI, forkhead box O3, inositol polyphosphate-4-phosphatase, LcK tyrosine kinase, phospho-PEA15 (Ser116), phospho-PRAS40, Rad51, and phospho-S6 (Ser240-244) ( 91 ).
The expression of maspin was also described as a marker for early recurrence in stage IV colorectal cancer. Snoeren et al. ( 92 ) analyzed tumor tissue samples from five stage IV patients with early recurrence (<6 months) and five patients with prolonged time to recurrence (>24 months) through mass spectrometry with subsequent validation by Western blotting. They demonstrated that maspin was differentially expressed in stage IV colorectal cancer patients with early and late recurrence after surgery for colorectal liver metastases ( 92 ).
Despite the cited potential prognostic biomarkers ( Table 3 ), carcinoembryonic antigen remains the only established protein biomarker in clinical practice to determine prognosis. The identification, validation, and translation of new prognostic biomarkers is important to fill the presented gaps in our knowledge, such as the prediction of nodal status, distance metastasis, and postsurgical recurrence.
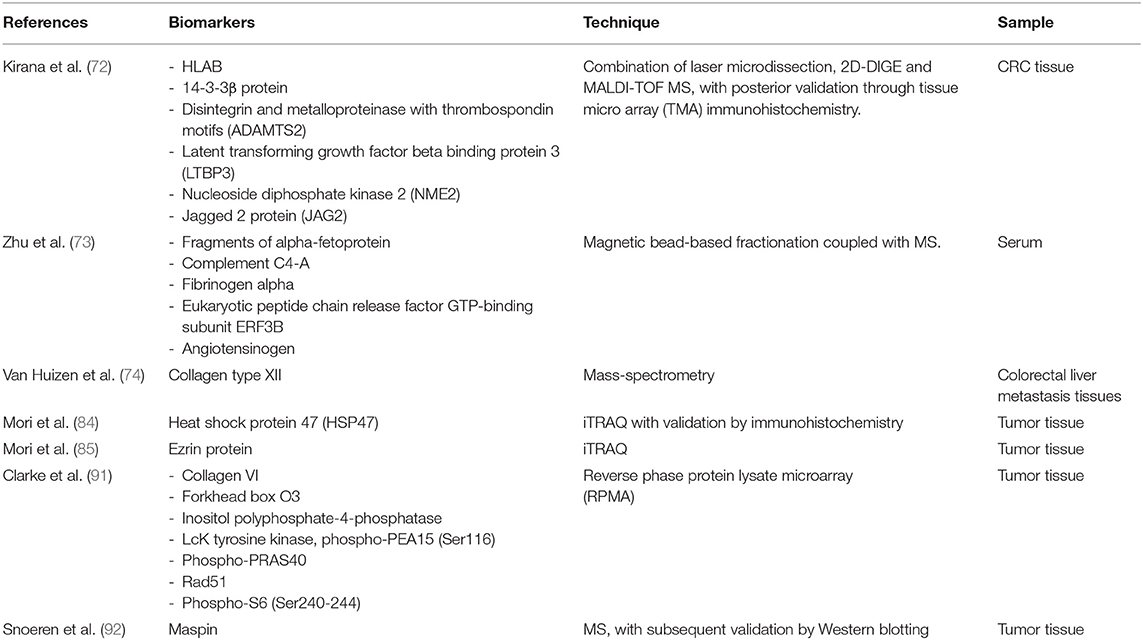
Table 3 . Examples of candidate prognostic biomarkers.
The approach to patients with colorectal cancer has been dramatically updated recently thanks to a better understanding of the process of carcinogenesis and advances in the field of genetics. The determination of KRAS, BRAF, and MSI status has become an indispensable step in therapeutic planning, especially in patients with metastatic disease. Furthermore, the emergence of immunotherapy and the increasing use of liquid biopsies have extended the possibilities in the decision-making process toward personalized medicine. However, even with these advances, there is a lack of biomarkers that can guide the early diagnosis or the targeted treatment, prognosis, and surveillance of patients with colorectal cancer. Despite the improvements in MS technologies and the large number of proteomics-based studies to find biomarkers, this approach is not mainstream, especially regarding the management of colorectal cancer patients. This difficulty in translating protein markers into clinical practice is probably due to the small sample sizes of studies and to the heterogeneity in the processes of sample obtainment, preparation, and storage. These factors, coupled with complexities of data analysis and interpretation of proteomic approaches, result in poor reproducibility of the studies. However, the greatest limitation is related to the absence of validation of the possible biomarkers in large cohorts, comparing the data with the current methods of diagnosis, prediction, and prognosis. In this scenario, translational proteomics remains a powerful and promising tool for the discovery of biomarkers that can lead to important changes in the management of patients with colorectal cancer. Probably, the key to personalized medicine in colorectal cancer relies on studies that can integrate genomic, transcriptomic, and proteomic data, from a multiomics point of view, in the search for a biomarker panel that combines strong clinical data and accurate molecular findings.
Author Contributions
BA has contributed to the drafting, writing, conceptualizing, and final revision of the manuscript. GB and IC have contributed to the writing, organizing of data and tables, and reference revision. MM has contributed to early drafting, writing, and reference surveys. PO has contributed to the conceptualizing of the manuscript. AM has contributed to the conceptualizing, writing, final revision, and supervision of the manuscript. All authors approved the review final form.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the team of researchers from the Laboratory of Biochemistry and Protein Chemistry (LBQP), Institute of Biology, Department of Cell Biology, University of Brasilia, Brazil. We greatly thank and acknowledge Ms. Raphaela Menezes de Oliveira for her support in improving this work. We also thank the members of the Colorectal Surgery Department from the University of Brasilia.
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
CrossRef Full Text | Google Scholar
2. Arvelo F, Sojo F, Cotte C. Biology of colorectal cancer. Ecancermedicalscience. (2015) 9:520. doi: 10.3332/ecancer.2015.520
PubMed Abstract | CrossRef Full Text | Google Scholar
3. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. (1990) 61:759–67. doi: 10.1016/0092-8674(90)90186-I
4. Ang CS, Baker MS, Nice EC. Mass spectrometry-based analysis for the discovery and validation of potential colorectal cancer stool biomarkers. Methods Enzymol. 586:247–74. doi: 10.1016/bs.mie.2016.10.019
5. Rutka M, Bor R, Bálint A, Fábián A, Milassin Á, Nagy F, et al. Diagnostic accuracy of five different fecal markers for the detection of precancerous and cancerous lesions of the colorectum. Mediat Inflammation. (2016) 2016:1–6. doi: 10.1155/2016/2492081
6. Brenner H, Chang-Claude J, Seiler CM, Rickert A, Hoffmeister M. Protection from colorectal cancer after colonoscopy: a population-based, case-control study. Ann Internal Med. (2011) 154:22–30. doi: 10.7326/0003-4819-154-1-201101040-00004
7. DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. (2014) 64:252–71. doi: 10.3322/caac.21235
8. Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. (2018) 68:250–81. doi: 10.3322/caac.21457
9. Al Bandar MH, Kim NK. Current status and future perspectives on treatment of liver metastasis in colorectal cancer. Oncol Rep. (2017) 37:2553–64. doi: 10.3892/or.2017.5531
10. Feo L, Polcino M, Nash GM. Resection of the primary tumor in stage IV colorectal cancer. Surg Clin N Am. (2017) 97:657–69. doi: 10.1016/j.suc.2017.01.012
11. Bailey JR, Aggarwal A, Imperiale TF. Colorectal cancer screening: stool DNA and other noninvasive modalities. Gut Liver. (2016) 10:204. doi: 10.5009/gnl15420
12. Coghlin C, Murray GI. Biomarkers of colorectal cancer: Recent advances and future challenges. Proteomics Clin Appl. (2015) 9:64–71. doi: 10.1002/prca.201400082
13. de Wit M, Fijneman RJA, Verheul HMW, Meijer GA, Jimenez CR. Proteomics in colorectal cancer translational research: biomarker discovery for clinical applications. Clin Biochem. (2013) 46:466–79. doi: 10.1016/j.clinbiochem.2012.10.039
14. Álvarez-Chaver P, Otero-Estévez O, de la Cadena MP, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Proteomics for discovery of candidate colorectal cancer biomarkers. World J Gastroenterol. (2014) 20:3804–24. doi: 10.3748/wjg.v20.i14.3804
15. Ma H, Chen G, Guo M. Mass spectrometry based translational proteomics for biomarker discovery and application in colorectal cancer. Proteomics Clin Appl. (2016) 10:503–15. doi: 10.1002/prca.201500082
16. Wang K, Huang C, Nice EC. Proteomics, genomics and transcriptomics: their emerging roles in the discovery and validation of colorectal cancer biomarkers. Expert Rev Proteomics. (2014) 11:179–205. doi: 10.1586/14789450.2014.894466
17. Langan RC, Mullinax JE, Raiji MT, Upham T, Summers T, Stojadinovic A, et al. Colorectal cancer biomarkers and the potential role of cancer stem cells. J Cancer. (2013) 4:241–50. doi: 10.7150/jca.5832
18. Lee PY, Chin S-F, Low TY, Jamal R. Probing the colorectal cancer proteome for biomarkers: current status and perspectives. J Proteomics. (2018) 187:93–105. doi: 10.1016/j.jprot.2018.06.014
19. Bhalla A, Zulfiqar M, Bluth MH. Molecular diagnostics in colorectal carcinoma. Clin Lab Med. (2018) 38:311–42. doi: 10.1016/j.cll.2018.02.008
20. Fernandes Messias MC, Mecatti GC, Figueiredo Angolini CF, Eberlin MN, Credidio L, Real Martinez CA, et al. Plasma lipidomic signature of rectal adenocarcinoma reveals potential biomarkers. Front Oncol. (2018) 7:325. doi: 10.3389/fonc.2017.00325
21. Perttula K, Schiffman C, Edmands WMB, Petrick L, Grigoryan H, Cai X, et al. Untargeted lipidomic features associated with colorectal cancer in a prospective cohort. BMC Cancer. (2018) 18:996. doi: 10.1186/s12885-018-4894-4
22. Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. Lipids Health Dis. (2019) 18:29. doi: 10.1186/s12944-019-0977-8
23. Quesada-Calvo F, Massot C, Bertrand V, Longuespée R, Blétard N, Somja J, et al. OLFM4, KNG1 and Sec24C identified by proteomics and immunohistochemistry as potential markers of early colorectal cancer stages. Clin Proteomics. (2017) 14:9. doi: 10.1186/s12014-017-9143-3
24. Alnabulsi A, Murray GI. Proteomics for early detection of colorectal cancer: recent updates. Expert Rev Proteomics. (2018) 15:55–63. doi: 10.1080/14789450.2018.1396893
25. Califf RM. Biomarker definitions and their applications. Exp Biol Med. (2018) 243:213–21. doi: 10.1177/1535370217750088
26. Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. (2012) 366:687–96. doi: 10.1056/NEJMoa1100370
27. Liang PS, Dominitz JA. Colorectal cancer screening. Med Clin N Am. (2019) 103:111–23. doi: 10.1016/j.mcna.2018.08.010
28. Church J. Complications of colonoscopy. Gastroenterol Clin N Am. (2013) 42:639–57. doi: 10.1016/j.gtc.2013.05.003
29. Alexander F, Weller D. E valuation of the UK Colorectal Cancer Screening Pilot . Final Report. The UK CRC Screening Pilot Evaluation Team.
Google Scholar
30. Logan RFA, Patnick J, Nickerson C, Coleman L, Rutter MD, von Wagner C. Outcomes of the Bowel Cancer Screening Programme (BCSP) in England after the first 1 million tests. Gut. (2012) 61:1439–46. doi: 10.1136/gutjnl-2011-300843
31. McLachlan S-A, Clements A, Austoker J. Patients' experiences and reported barriers to colonoscopy in the screening context—A systematic review of the literature. Patient Educ Counsel. (2012) 86:137–46. doi: 10.1016/j.pec.2011.04.010
32. Plumb AA, Ghanouni A, Rainbow S, Djedovic N, Marshall S, Stein J, et al. Patient factors associated with non-attendance at colonoscopy after a positive screening faecal occult blood test. J Med Screen. (2017) 24:12–9. doi: 10.1177/0969141316645629
33. Navarro M, Nicolas A, Ferrandez A, Lanas A. Colorectal cancer population screening programs worldwide in 2016: an update. World J Gastroenterol. (2017) 23:3632. doi: 10.3748/wjg.v23.i20.3632
34. Kościelniak-Merak B, Radosavljević B, Zajac A, Tomasik PJ. Faecal occult blood point-of-care tests. J Gastrointest Cancer. (2018) 49:402–5. doi: 10.1007/s12029-018-0169-1
35. Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol. (2017) 23:5086. doi: 10.3748/wjg.v23.i28.5086
36. Ghazanfar S, Fatima I, Aslam M, Musharraf SG, Sherman NE, Moskaluk C, et al. Identification of actin beta-like 2 (ACTBL2) as novel, upregulated protein in colorectal cancer. J Proteomics. (2017) 152:33–40. doi: 10.1016/j.jprot.2016.10.011
37. Hao J-J, Zhi X, Wang Y, Zhang Z, Hao Z, Ye R, et al. Comprehensive proteomic characterization of the human colorectal carcinoma reveals signature proteins and perturbed pathways. Sci Rep. (2017) 7:42436. doi: 10.1038/srep42436
38. Yamamoto T, Kudo M, Peng W-X, Takata H, Takakura H, Teduka K, et al. Identification of aldolase A as a potential diagnostic biomarker for colorectal cancer based on proteomic analysis using formalin-fixed paraffin-embedded tissue. Tumor Biol. (2016) 37:13595–606. doi: 10.1007/s13277-016-5275-8
39. Ganepola GA. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J Gastrointest Oncol. (2014) 6:83. doi: 10.4251/wjgo.v6.i4.83
40. Ivancic MM, Megna BW, Sverchkov Y, Craven M, Reichelderfer M, Pickhardt PJ, et al. Noninvasive detection of colorectal carcinomas using serum protein biomarkers. J Surg Res. (2020) 246:160–9. doi: 10.1016/j.jss.2019.08.004
41. Bhardwaj M, Gies A, Weigl K, Tikk K, Benner A, Schrotz-King P, et al. Evaluation and validation of plasma proteins using two different protein detection methods for early detection of colorectal cancer. Cancers. (2019) 11:1426. doi: 10.3390/cancers11101426
42. Yu J, Zhai X, Li X, Zhong C, Guo C, Yang F, Yuan Y, Zheng S. Identification of MST1 as a potential early detection biomarker for colorectal cancer through a proteomic approach. Sci Rep. (2017) 7:14265. doi: 10.1038/s41598-017-14539-x
43. Fan N-J, Chen H-M, Song W, Zhang Z-Y, Zhang M-D, Feng L-Y, et al. Macrophage mannose receptor 1 and S100A9 were identified as serum diagnostic biomarkers for colorectal cancer through a label-free quantitative proteomic analysis. Cancer Biomark. (2016) 16:235–43. doi: 10.3233/CBM-150560
44. Peltier J, Roperch J-P, Audebert S, Borg J-P, Camoin L. Quantitative proteomic analysis exploring progression of colorectal cancer: modulation of the serpin family. J Proteomics. (2016) 148:139–48. doi: 10.1016/j.jprot.2016.07.031
45. Alnabulsi A, Murray GI. Integrative analysis of the colorectal cancer proteome: potential clinical impact. Expert Rev Proteomics. (2016) 13:917–27. doi: 10.1080/14789450.2016.1233062
46. Chauvin A, Boisvert F-M. Clinical proteomics in colorectal cancer, a promising tool for improving personalised medicine. Proteomes. (2018) 6:49. doi: 10.3390/proteomes6040049
47. Wang J, Mouradov D, Wang X, Jorissen RN, Chambers MC, Zimmerman LJ, et al. Colorectal cancer cell line proteomes are representative of primary tumors and predict drug sensitivity. Gastroenterology. (2017) 153:1082–95. doi: 10.1053/j.gastro.2017.06.008
48. Guo J, Zhu C, Yang K, Li J, Du N, Zong M, et al. Poly(C)-binding protein 1 mediates drug resistance in colorectal cancer. Oncotarget. (2017) 8:13312–9. doi: 10.18632/oncotarget.14516
49. Martin P, Noonan S, Mullen MP, Scaife C, Tosetto M, Nolan B, et al. Predicting response to vascular endothelial growth factor inhibitor and chemotherapy in metastatic colorectal cancer. BMC Cancer. (2014) 14:887. doi: 10.1186/1471-2407-14-887
50. Katsila T, Juliachs M, Gregori J, Macarulla T, Villarreal L, Bardelli A, et al. Circulating pEGFR is a candidate response biomarker of cetuximab therapy in colorectal cancer. Clin Cancer Res. (2014) 20:6346–56. doi: 10.1158/1078-0432.CCR-14-0361
51. Passardi A, Canale M, Valgiusti M, Ulivi P. Immune checkpoints as a target for colorectal cancer treatment. Int J Mol Sci. (2017) 18:1324. doi: 10.3390/ijms18061324
52. Overman MJ, Lonardi S, Leone F, McDermott RS, Morse MA, Wong KYM, et al. Nivolumab in patients with DNA mismatch repair deficient/microsatellite instability high metastatic colorectal cancer: Update from CheckMate 142. J Clin Oncol. (2017) 35:519. doi: 10.1200/JCO.2017.35.4_suppl.519
53. Diaz LA, Marabelle A, Delord J-P, Shapira-Frommer R, Geva R, Peled N, et al. Pembrolizumab therapy for microsatellite instability high (MSI-H) colorectal cancer (CRC) and non-CRC. J Clin Oncol. (2017) 35:3071. doi: 10.1200/JCO.2017.35.15_suppl.3071
54. Bendell JC, Powderly JD, Lieu CH, Eckhardt SG, Hurwitz H, Hochster HS, et al. Safety and efficacy of MPDL3280A (anti-PDL1) in combination with bevacizumab (bev) and/or FOLFOX in patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol. (2015) 33:704. doi: 10.1200/jco.2015.33.3_suppl.704
55. Yaghoubi N, Soltani A, Ghazvini K, Hassanian SM, Hashemy SI. PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed Pharmacother. (2019) 110:312–8. doi: 10.1016/j.biopha.2018.11.105
56. Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. (2015) 5:43–51. doi: 10.1158/2159-8290.CD-14-0863
57. Battaglin F, Naseem M, Lenz H-J, Salem ME. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol. (2018) 16:735–45.
PubMed Abstract | Google Scholar
58. Oh HJ, Kim JH, Lee TH, Park HE, Bae JM, Lee HS, Kang GH. Dominant high expression of wild-type HSP110 defines a poor prognostic subgroup of colorectal carcinomas with microsatellite instability: a whole-section immunohistochemical analysis. APMIS. (2017) 125:1076–83. doi: 10.1111/apm.12770
59. Janikovits J, Müller M, Krzykalla J, Körner S, Echterdiek F, Lahrmann B, et al. High numbers of PDCD1 (PD-1)-positive T cells and B2M mutations in microsatellite-unstable colorectal cancer. Oncoimmunology. (2018) 7:e1390640. doi: 10.1080/2162402X.2017.1390640
60. Yang Q, Roehrl MH, Wang JY. Proteomic profiling of antibody-inducing immunogens in tumor tissue identifies PSMA1, LAP3, ANXA3, and maspin as colon cancer markers. Oncotarget. (2018) 9:3996–4019. doi: 10.18632/oncotarget.23583
61. Yang Q, Bavi P, Wang JY, Roehrl MH. Immuno-proteomic discovery of tumor tissue autoantigens identifies olfactomedin 4, CD11b, and integrin alpha-2 as markers of colorectal cancer with liver metastases. J Proteomics. (2017) 168:53–65. doi: 10.1016/j.jprot.2017.06.021
62. Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen Y-J, Ciombor KK, et al. NCCN guidelines insights: colon cancer, version 2.2018. J Natl Comp Cancer Netw. (2018) 16:359–69. doi: 10.6004/jnccn.2018.0021
63. Habr-Gama A, Perez RO, Nadalin W, Sabbaga J, Ribeiro U, Silva e Sousa AH, et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy. Trans Meet Am Surg Assoc. (2004) 122:309–16. doi: 10.1097/01.sla.0000141194.27992.32
64. Chauvin A, Wang C-S, Geha S, Garde-Granger P, Mathieu A-A, Lacasse V, et al. The response to neoadjuvant chemoradiotherapy with 5-fluorouracil in locally advanced rectal cancer patients: a predictive proteomic signature. Clin Proteomics. (2018) 15:16. doi: 10.1186/s12014-018-9192-2
65. Oldenhuis CNAM, Oosting SF, Gietema JA, de Vries EGE. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. (2008) 44:946–53. doi: 10.1016/j.ejca.2008.03.006
66. Weiser MR. AJCC 8th edition: colorectal cancer. Ann Surg Oncol. (2018) 25:1454–5. doi: 10.1245/s10434-018-6462-1
67. Gonzalez-Pons M, Cruz-Correa M. Colorectal cancer biomarkers: where are we now? BioMed Res Int. (2015) 2015:1–14. doi: 10.1155/2015/149014
68. Campos-da-Paz M, Dórea JG, Galdino AS, Lacava ZGM, de Fatima Menezes Almeida Santos M. Carcinoembryonic Antigen (CEA) and hepatic metastasis in colorectal cancer: update on biomarker for clinical and biotechnological approaches. Recent Patents Biotechnol. (2018) 12:269–79. doi: 10.2174/1872208312666180731104244
69. O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new american joint committee on cancer sixth edition staging. J Natl Cancer Inst. (2004) 96:1420–5. doi: 10.1093/jnci/djh275
70. Hugen N, van de Velde CJH, de Wilt JHW, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. (2014) 25:651–7. doi: 10.1093/annonc/mdt591
71. Marfà S, Marti J, Reyes A, Casals G, Fernández-Varo G, Carvajal S, et al. Metastatic tissue proteomic profiling predicts 5-year outcomes in patients with colorectal liver metastases. Transl Oncol. (2016) 9:445–52. doi: 10.1016/j.tranon.2016.08.002
72. Kirana C, Peng L, Miller R, Keating JP, Glenn C, Shi H, et al. Combination of laser microdissection, 2D-DIGE and MALDI-TOF MS to identify protein biomarkers to predict colorectal cancer spread. Clin Proteomics. (2019) 16:3. doi: 10.1186/s12014-019-9223-7
73. Zhu D, Zhong Y, Wu H, Ye L, Wang J, Li Y, et al. Predicting metachronous liver metastasis from colorectal cancer using serum proteomic fingerprinting. J Surg Res. (2013) 184:861–6. doi: 10.1016/j.jss.2013.04.065
74. van Huizen NA, Coebergh van den Braak RRJ, Doukas M, Dekker LJM, IJzermans JNM, et al. Up-regulation of collagen proteins in colorectal liver metastasis compared with normal liver tissue. J Biol Chem. (2019) 294:281–9. doi: 10.1074/jbc.RA118.005087
75. Lalmahomed Z, Broker M, Roest H, Van Huizen N, Dekker L, Calame W, et al. P-0284 * collagen peptides in urine: a new promising biomarker for the detection of colorectal liver metastases. Ann Oncol. (2013) 24:iv116. doi: 10.1093/annonc/mdt203.282
76. Lalmahomed ZS, Bröker ME, van Huizen NA, Coebergh van den Braak RRJ, Dekker LJ, Rizopoulos D, et al. Hydroxylated collagen peptide in urine as biomarker for detecting colorectal liver metastases. Am J Cancer Res. (2016) 6:321–30. doi: 10.1200/JCO.2016.34.15_suppl.e15081
77. Altobelli E, Angeletti PM, Latella G. Role of urinary biomarkers in the diagnosis of adenoma and colorectal cancer: a systematic review and meta-analysis. J Cancer. (2016) 7:1984–2004. doi: 10.7150/jca.16244
78. Colbert Maresso K, Vilar E, Hawk ET. Urinary PGE-M in colorectal cancer: predicting more than risk? Cancer Prevent Res. (2014) 7:969–72. doi: 10.1158/1940-6207.CAPR-14-0215
79. Bezawada N, Song M, Wu K, Mehta RS, Milne GL, Ogino S, et al. Urinary PGE-M levels are associated with risk of colorectal adenomas and chemopreventive response to anti-inflammatory drugs. Cancer Prevent Res. (2014) 7:758–65. doi: 10.1158/1940-6207.CAPR-14-0120
80. Shrubsole MJ, Cai Q, Wen W, Milne G, Smalley WE, Chen Z, et al. Urinary prostaglandin E2 metabolite and risk for colorectal adenoma. Cancer Prevent Res. (2012) 5:336–42. doi: 10.1158/1940-6207.CAPR-11-0426
81. Johnson JC, Schmidt CR, Shrubsole MJ, Billheimer DD, Joshi PR, Morrow JD, et al. Urine PGE-M: a metabolite of prostaglandin E2 as a potential biomarker of advanced colorectal neoplasia. Clin Gastroenterol Hepatol. (2006) 4:1358–365. doi: 10.1016/j.cgh.2006.07.015
82. Ong MLH, Schofield JB. Assessment of lymph node involvement in colorectal cancer. World J Gastrointest Surg. (2016) 8:179. doi: 10.4240/wjgs.v8.i3.179
83. Low G, Tho LM, Leen E, Wiebe E, Kakumanu S, McDonald AC, et al. The role of imaging in the pre-operative staging and post-operative follow-up of rectal cancer. Surgeon. (2008) 6:222–31. doi: 10.1016/S1479-666X(08)80032-7
84. Mori K, Toiyama Y, Otake K, Fujikawa H, Saigusa S, Hiro J, et al. Proteomics analysis of differential protein expression identifies heat shock protein 47 as a predictive marker for lymph node metastasis in patients with colorectal cancer: HSP47 expression in CRC. Int J Cancer. (2017) 140:1425–35. doi: 10.1002/ijc.30557
85. Mori K, Toiyama Y, Otake K, Ide S, Imaoka H, Okigami M, et al. Successful identification of a predictive biomarker for lymph node metastasis in colorectal cancer using a proteomic approach. Oncotarget. (2017) 8:106935–47. doi: 10.18632/oncotarget.22149
86. Liang F, Wang Y, Shi L, Zhang J. Association of Ezrin expression with the progression and prognosis of gastrointestinal cancer: a meta-analysis. Oncotarget. (2017) 8:93186–95. doi: 10.18632/oncotarget.21473
87. Das V, Kalita J, Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacother. (2017) 87:8–19. doi: 10.1016/j.biopha.2016.12.064
88. Jörgren F, Nilbert M, Rambech E, Bendahl P-O, Lindmark G. Ezrin expression in rectal cancer predicts time to development of local recurrence. Int J Colorectal Dis. (2012) 27:893–9. doi: 10.1007/s00384-011-1397-z
89. Patara M, Santos EMM, Coudry R de A, Soares FA, Ferreira FO, Rossi BM. Ezrin expression as a prognostic marker in colorectal adenocarcinoma. Pathol Oncol Res. (2011) 17:827–33. doi: 10.1007/s12253-011-9389-4
90. Duineveld LAM, van Asselt KM, Bemelman WA, Smits AB, Tanis PJ, van Weert HCPM, et al. Symptomatic and asymptomatic colon cancer recurrence: a multicenter cohort study. Ann Family Med. (2016) 14:215–20. doi: 10.1370/afm.1919
91. Clarke CN, Lee MS, Wei W, Manyam G, Jiang ZQ, Lu Y, et al. Proteomic features of colorectal cancer identify tumor subtypes independent of oncogenic mutations and independently predict relapse-free survival. Ann Surg Oncol. (2017) 24:4051–8. doi: 10.1245/s10434-017-6054-5
92. Snoeren N, Emmink BL, Koerkamp MJG, van Hooff SR, Goos JACM, van Houdt WJ, et al. Maspin is a marker for early recurrence in primary stage III and IV colorectal cancer. Brit J Cancer. (2013) 109:1636–47. doi: 10.1038/bjc.2013.489
Keywords: colorectal cancer, biomarkers, translational research, proteomics, mass spectrometry
Citation: Alves Martins BA, de Bulhões GF, Cavalcanti IN, Martins MM, de Oliveira PG and Martins AMA (2019) Biomarkers in Colorectal Cancer: The Role of Translational Proteomics Research. Front. Oncol. 9:1284. doi: 10.3389/fonc.2019.01284
Received: 02 June 2019; Accepted: 05 November 2019; Published: 27 November 2019.
Reviewed by:
Copyright © 2019 Alves Martins, de Bulhões, Cavalcanti, Martins, de Oliveira and Martins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bruno Augusto Alves Martins, brunomartins.coloprocto@gmail.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
- Open access
- Published: 14 April 2022
Diagnostic value of tumor markers in identifying favorable or unfavorable subsets in patients with cancer of unknown primary: a retrospective study
- Shigemasa Takamizawa 1 ,
- Tatsunori Shimoi 1 ,
- Masayuki Yoshida 2 ,
- Momoko Tokura 1 ,
- Shu Yazaki 1 ,
- Chiharu Mizoguchi 1 ,
- Ayumi Saito 1 ,
- Shosuke Kita 1 ,
- Kasumi Yamamoto 1 ,
- Yuki Kojima 1 ,
- Hitomi Sumiyoshi-Okuma 1 ,
- Tadaaki Nishikawa 1 ,
- Emi Noguchi 1 ,
- Kazuki Sudo 1 &
- Kan Yonemori 1
BMC Cancer volume 22 , Article number: 412 ( 2022 ) Cite this article
2729 Accesses
2 Citations
1 Altmetric
Metrics details
Routine measurement of tumor markers is not recommended in daily clinical practice for patients with cancer of unknown primary (CUP). We evaluated the diagnostic value of tumor markers in identifying favorable or unfavorable subsets in patients with CUP.
We retrospectively reviewed the medical records of patients who were diagnosed with CUP between October 2010 and July 2015 at the National Cancer Center Hospital. The tumor markers of the patients were examined, including squamous cell carcinoma antigen, cytokeratin fraction, carcinoembryonic antigen, sialyl Lewis X, neuron-specific enolase, pro-gastrin-releasing peptide, α-fetoprotein, protein induced by vitamin K absence or antagonist II, prostate-specific antigen, soluble interleukin-2 receptor, carbohydrate antigen 19–9, cancer antigen 125, cancer antigen 15–3, NCC-ST-439 (ST439), elastase-1, human chorionic gonadotropin, and sialyl-Tn (STN).
Among 199 patients with suspected CUP, 90 were diagnosed with confirmed CUP (12 in the favorable subset and 78 in the unfavorable subset). No tumor markers showed 100% sensitivity for unfavorable subsets. ST439 ( p = 0.03) and STN ( p = 0.049) showed 100% specificity for unfavorable subsets.
Conclusions
For patients with suspected CUP who show elevated ST439 or STN levels, the treatment strategy should be based on the premise that the patient is likely to be placed in the unfavorable subset.
Peer Review reports
Cancer of unknown primary (CUP) is defined as a cancer lacking any detectable primary site after full evaluation. Only metastatic sites are histologically confirmed. CUP is a rare malignancy, accounting for approximately 3%–5% of all newly diagnosed patients with malignancies [ 1 ]. In addition, some are found to be non-cancerous during a thorough examination [ 2 ]. Approximately 20% of patients with CUP have a favorable prognosis [ 1 ]. This patient group includes men with adenocarcinoma of bone metastases and elevated prostate-specific antigen (PSA), women with papillary adenocarcinoma of the peritoneal cavity, women with adenocarcinoma involving the axillary lymph nodes, patients with poorly differentiated carcinoma with midline distribution, patients with well-differentiated neuroendocrine tumors or poorly differentiated neuroendocrine carcinomas, patients with squamous cell carcinoma involving cervical lymph nodes, patients with adenocarcinoma with a colon cancer profile, and patients with squamous cell carcinoma of isolated inguinal adenopathy [ 1 ]. These patients should be identified at initial evaluation and receive specific therapy to extend the prognosis.
Approximately 80% of patients with CUP do not have any favorable subsets, and the prognosis of these patients is worse [ 1 ]. The median survival time of these patients is only 6–7 months [ 1 ], and a standard of care for this patient group is absent [ 3 ]. Although most patients with unfavorable subsets are treated based on suspected tissue-of-origin, there is no survival advantage compared with empiric platinum-based combination chemotherapy [ 4 ]. A previous report analyzed 93 patients who received platinum-based combination chemotherapy, and the response rate was 39.8% [ 5 ]. A meta-analysis has shown that no type of chemotherapy has been proven to lengthen survival time [ 6 ].
Evaluation of tumor markers is useful for diagnosis and the reduction of inappropriate diagnostic tests for patients with suspected malignancy [ 7 ]. Although patients with CUP commonly overexpress several tumor markers, the diagnostic, predictive, and prognostic utilities are unexplained. Routine measurement of tumor markers for patients with CUP is not recommended in daily clinical practice [ 8 ].
Tumor markers are not recommended for finding the primary site of CUP, except in limited situations. The guidelines published by the European Society for Medical Oncology mention that useful tumor markers for diagnosing the primary tumor site include human chorionic gonadotropin (hCG) and α-fetoprotein (AFP) in patients with poorly differentiated carcinoma of midline distribution for germ-cell tumors, PSA in men with bone metastases for prostate cancer, cancer antigen 125 (CA125) in women with primary peritoneal serous adenocarcinoma for ovarian, fallopian tube, and peritoneal cancers, and thyroglobulin for differentiated thyroid cancer [ 1 , 8 , 9 , 10 , 11 , 12 , 13 ].
Squamous cell carcinoma antigen is a marker that is elevated in squamous cell carcinomas, such as head and neck, esophageal, and uterine cervical cancers [ 14 ]. Cytokeratin fraction (cytokeratin 19 fragment) is elevated in non-small cell lung cancers [ 15 ]. Carcinoembryonic antigen, present in the fetal digestive cells, is elevated in gastric, colorectal, and other cancers of the digestive system [ 16 ]. Sialyl Lewis X is a polymeric glycoprotein elevated in lung, ovarian, and pancreatic cancers [ 17 ]. Neuron-specific enolase increases with the tumorigenesis in neuroendocrine cells, such as in small-cell lung cancer and neuroblastoma [ 18 ]. Pro-gastrin-releasing peptide is a gastrointestinal hormone; it is a marker for small-cell lung cancer [ 19 ]. Protein induced by vitamin K absence or antagonist II is precursor of the coagulation factor prothrombin; it is a marker for hepatocellular carcinoma [ 20 ]. Soluble interleukin-2 receptor is the alpha chain of interleukin 2 receptor, which exists in the free-form in blood; it is elevated in lymphoid malignancies such as non-Hodgkin's lymphoma, adult T-cell lymphoma/leukemia, and acute lymphocytic leukemia [ 21 ]. Carbohydrate antigen 19–9 is a cell surface glycoprotein complex; it is elevated in gastrointestinal cancers, such as pancreatic, gallbladder, and bile duct cancers [ 22 ]. Cancer antigen 15–3 is a mucin-type glycoprotein, which is elevated in breast cancers [ 23 ]. NCC-ST 439 is a mucin-type glycoprotein that is elevated in breast and gastrointestinal cancers [ 24 ]. Elastase-1 is a proteolytic enzyme; it is a marker for pancreatic cancer [ 25 ]. Sialyl-Tn is a sugar chain antigen; it is elevated in ovarian and gastrointestinal cancers [ 26 ]. However, these tumor markers are not recommended for identifying the primary site of CUP.
The diagnostic evaluation of patients with CUP takes time, sometimes up to several months. In addition, deciding whether a subset is favorable or not must be carefully considered because it has a great impact on the treatment selection and prognosis. Only a few studies have examined whether tumor markers can be used to classify subsets. If tumor markers could be used to classify favorable and unfavorable subsets, then the best treatment option could be more quickly recommended to patients with CUP. Identifying favorable subsets during the initial evaluation can lead to appropriate treatment and prolonged survival. We evaluated the diagnostic value of tumor markers that are routinely used in our hospital for identifying favorable or unfavorable subsets in patients with CUP.
Study cohort
We retrospectively reviewed the medical records of patients who were diagnosed with CUP at National Cancer Center Hospital (NCCH) (Tokyo, Japan) between October 2010 and July 2015. This single-institution medical record-based retrospective observational study was approved by the Institutional Review Board of NCCH (NCCH 2012–335), which waived the requirement for informed consent. Patient registration was based on an opt-out model. The study was conducted according to the principles of the Declaration of Helsinki.
Diagnosis of CUP
Since our facility is a cancer-specialized hospital, most patients with suspected cancer were referred to us before they had undergone adequate examination. Patients then underwent a fundamental workup and additional focused imaging based on their cancer distribution and histopathology. Examinations were performed according to the guidelines of the European Society for Medical Oncology and the Japanese Society of Medical Oncology [ 12 ].
Patients were evaluated through an initial workup, including physical examination, laboratory studies (a complete blood count, urinalysis, basic serum chemistries, and tumor marker analysis), and imaging procedures (computed tomography scan or magnetic resonance imaging (MRI) of the chest, abdomen, and pelvis). The selected women were evaluated with a pelvic examination by a gynecologist or mammography and breast MRIs to look for breast lesions. For selected men or women, an examination of the prostate or urinary tract by a urologist was completed to look for urinary tract lesions.
The diagnosis of CUP was confirmed when the primary site of cancer was unknown after these initial workups, based on the consensus of medical oncology specialists.

Tumor markers
The patients were evaluated for the following tumor markers: squamous cell carcinoma antigen (cut-off: 1.5 ng/ml), cytokeratin 19 fragment (cut-off: 2.2 ng/ml), carcinoembryonic antigen (cut-off: 5.0 ng/ml), sialyl Lewis X (cut-off: 38.0 U/ml), neuron-specific enolase (cut-off: 15.0 ng/ml), pro-gastrin-releasing peptide (cut-off: 81.0 pg/ml), AFP (cut-off: 10.0 ng/ml), protein induced by vitamin K absence or antagonist II (PIVKA-II) (cut-off: 40 mAU/ml), PSA (cut-off: 2.7 ng/ml), soluble interleukin-2 receptor (sIL-2R) (cut-off: 587 U/ml), carbohydrate antigen 19–9 (cut-off: 37 U/ml), CA125 (cut-off: 35 U/ml), cancer antigen 15–3 (cut-off: 28 U/ml), NCC-ST 439 (ST439) (cut-off: 4.5 U/ml), elastase-1 (cut-off: 300 ng/dl), hCG (cut-off: 3.0 mIU/ml), and sialyl-Tn (STN) (cut-off: 45.0 U/ml). These cut-off values were based on the facility standard.
Definition of favorable and unfavorable subsets
The following patient populations were placed in the favorable subset: men with adenocarcinoma who have bone metastases and elevated PSA; women with adenocarcinoma who have peritoneal carcinomatosis; women with adenocarcinoma who have axillary lymph node metastases; patients with poorly differentiated carcinoma with midline distribution; patients with well-differentiated neuroendocrine tumors or poorly differentiated neuroendocrine carcinomas; patients with squamous cell carcinoma involving the cervical nodes; patients with a colon cancer profile; and patients with squamous cell carcinoma of isolated inguinal adenopathy [ 12 , 13 ]. Patients without any of these factors were placed in the unfavorable subset.
Statistical analyses
Univariate analyses were performed to evaluate the correlation between tumor markers and favorable or unfavorable subsets. All statistical analyses were performed using JMP software (version 14.3.0 for Windows; SAS Institute Japan Inc., Cary, NC, USA), and results were considered significant with a two-sided p -value of < 0.05.
Patient characteristics
Between October 2010 and July 2015, 199 patients with suspected CUP were referred to the NCCH. Among them, 190 patients were examined via tumor markers, 100 were diagnosed with cancer of known primary site, and 90 were diagnosed with confirmed CUP (12 in the favorable subset and 78 in the unfavorable subset) (Fig. 1 ).
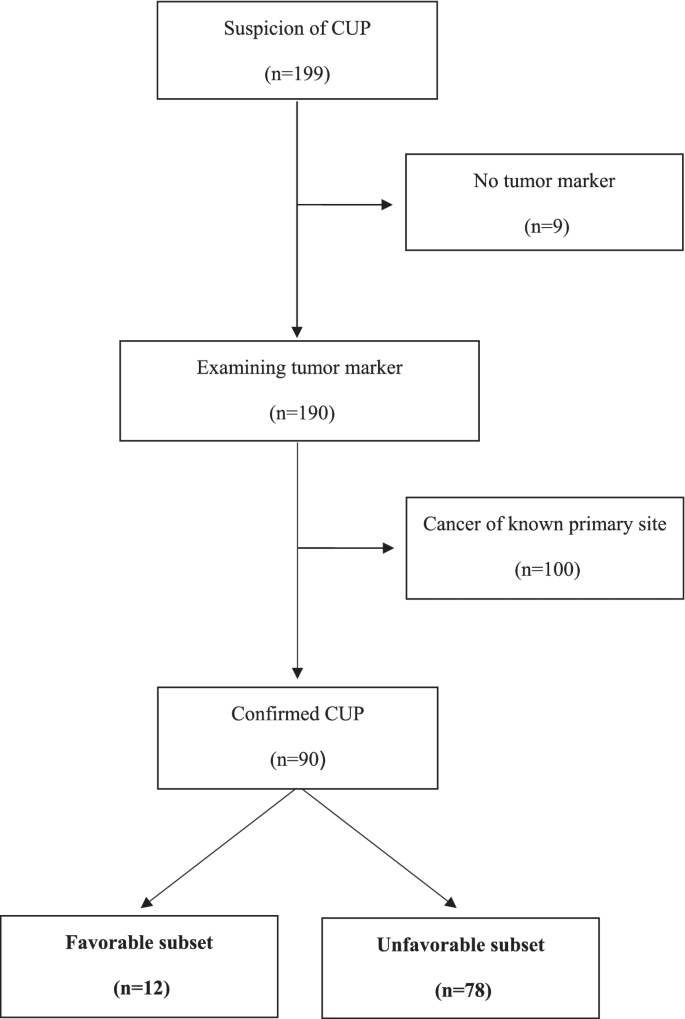
CONSORT diagram of patient selection
Median age was 68 years (range: 47–78) in the favorable subset and 66 years (range: 34–83) in the unfavorable subset. Females accounted for 83% of the favorable subset and 55% of the unfavorable subset. The Eastern Cooperative Oncology Group Performance Status (ECOG-PS) of most patients was 0 or 1. The estimated primary organs of 12 patients in the favorable subset were breast (5 patients), ovary/peritoneum (5 patients), and skin (2 patients). The characteristics of the patients are shown in Table 1 .
Sensitivity and specificity of tumor markers
No tumor markers showed 100% sensitivity for unfavorable subsets. However, PIVKA-II, PSA, ST439, elastase-1, and STN showed 100% specificity for unfavorable subsets. Among them, ST439 ( p = 0.03) and STN ( p = 0.049) showed a significant correlation between favorable and unfavorable subsets (Table 2 ).
Treatment regimen
Among the patients placed in the unfavorable subset, many were treated with drug therapy based on suspected tissue-of-origin, such as lung, based on the consensus of medical oncologists (Table 3 and 4 ). Patients within the normal range of ST439 were significantly more likely to receive drug therapy for ovarian cancer ( p = 0.004). Patients with elevated ST439 levels were significantly more likely to receive drug therapy for colorectal ( p = 0.036) or salivary gland cancer ( p = 0.031).
Despite tumor markers being easily accessible, their diagnostic ability for patients in unfavorable subsets had previously been unknown. Thus, we have evaluated tumor markers to identify patients in unfavorable subsets. ST439 and STN showed 100% specificity for patients in the unfavorable subset. No patients with elevated ST439 or STN above the reference value in the favorable subset were detected. In about 30% of the patients in the unfavorable subset, ST439 or STN was above the reference range. These results demonstrate that when ST439 or STN is elevated at the initial workup, a patient could be included in the unfavorable subset. In CUP treatment, the final diagnosis is not based soley on pathology, but on a combination of clinical factors. In addition, the standard of care for patients in unfavorable subsets is absent [ 3 ] and their prognosis is worse [ 1 ]. Therefore, it is necessary to differentiate between the favorable and unfavorable subsets within a limited time frame, such as one month [ 12 ]. Based on the findings from this study, routine evaluation of ST439 and STN could enable screening for treatment-ineffective subsets and prognostic estimation. This would enable identifying unfavorable subsets during the initial assessment for CUP. Refraining from aggressive treatments for patients in unfavorable subsets, who have a poor ECOG-PS, and early preparation for palliative care could improve the patients' quality of life. Evaluation of ST439 and STN at a patient’s first visit may help in the initial diagnosis of CUP in daily practice.
On the contrary, markers such as CA125, hCG, and PSA did not show a significant correlation between favorable and unfavorable subsets. This suggests that these markers are helpful when confirming favorable subsets with other clinical findings but are difficult to use alone for distinguishing between favorable and unfavorable subsets. The favorable subset included only five patients whose estimated primary organ was ovarian/peritoneal. There were no patients whose estimated primary organ was prostate or germ cell. A larger sample size is needed for further assessment. In addition, recent advances in histopathological examination of germ-cell tumors and malignant lymphomas suggest that anaplastic carcinoma of median development may not remain in a favorable subset as previously thought [ 27 ]. Therefore, the diagnostic abilities of tumor markers associated with malignant lymphomas (sIL-2R) and germ-cell tumors (hCG and AFP) may be limited given the current state of medicine.
An analysis comparing tumor markers and survival outcomes could not be carried out in this study because anticancer therapy was selected based on CUP histology or metastatic distribution and varied from patient to patient. We selected a primary site-directed treatment based on the suspected primary organ evaluated by a panel of oncologists. A previous report showed that patients with unfavorable subset CUP whose suspected primary organ was breast or ovary had higher response rates and a better prognosis compared with other unfavorable subsets [ 28 ].
This study has several limitations. It had a retrospective design and a relatively small sample size, with all data obtained from a single institution. In addition, the cut-off values were selected based on the facility standard. Whether these values are appropriate for distinguishing between favorable and unfavorable subsets of patients with CUP is unknown. Moreover, it is unclear why ST439 and STN can identify favorable or unfavorable subsets.
Additional research is needed regarding tumor markers that can identify favorable or unfavorable subsets in patients with CUP. Tumor markers can be utilized for the diagnosis of CUP in daily clinical practice.
We evaluated diagnostic value of tumor markers in identifying favorable or unfavorable subsets in patients with CUP. ST439 and STN showed 100% specificity for the unfavorable subset. If ST439 or STN is elevated in patients with CUP, they could be included in the unfavorable subset.
Availability of data and materials
The datasets generated during and analyzed during the current study are not publicly available due to protect patient privacy but are available from the corresponding author on reasonable request.
Abbreviations
α-Fetoprotein
Cancer antigen 125
- Cancer of unknown primary
Eastern cooperative oncology group-performance status
Human chorionic gonadotropin
Magnetic resonance imaging
Not applicable
National Cancer Center Hospital
Protein induced by vitamin K absence or antagonist II
prostate-specific antigen
Soluble interleukin-2 receptor
Sialyl Lewis X
Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379:1428–35.
Article PubMed Google Scholar
Sato J, Shimoi T, Shimomura A, Noguchi E, Kodaira M, Yunokawa M, et al. The incidence of nonmalignant diseases among patients with suspected carcinoma of unknown primary site. Intern Med. 2019;58:1423–8.
Article PubMed PubMed Central Google Scholar
Tomuleasa C, Zaharie F, Muresan MS, Pop L, Fekete Z, Dima D, et al. How to diagnose and treat a cancer of unknown primary site. J Gastrointestin Liver Dis. 2017;26:69–79.
Kato S, Alsafar A, Walavalkar V, Hainsworth J, Kurzrock R. Cancer of unknown primary in the molecular era. Trends Cancer. 2021;7:465–77.
Article CAS PubMed PubMed Central Google Scholar
Yonemori K, Ando M, Shibata T, Katsumata N, Matsumoto K, Yamanaka Y, et al. Tumor-marker analysis and verification of prognostic models in patients with cancer of unknown primary, receiving platinum-based combination chemotherapy. J Cancer Res Clin Oncol. 2006;132:635–42.
Article CAS PubMed Google Scholar
Golfinopoulos V, Pentheroudakis G, Salanti G, Nearchou AD, Ioannidis JP, Pavlidis N. Comparative survival with diverse chemotherapy regimens for cancer of unknown primary site: multiple-treatments meta-analysis. Cancer Treat Rev. 2009;35:570–3.
Molina R, Bosch X, Auge JM, Filella X, Escudero JM, Molina V, et al. Utility of serum tumor markers as an aid in the differential diagnosis of patients with clinical suspicion of cancer and in patients with cancer of unknown primary site. Tumour Biol. 2012;33:463–74.
Pavlidis N, Briasoulis E, Hainsworth J, Greco FA. Diagnostic and therapeutic management of cancer of an unknown primary. Eur J Cancer. 2003;39:1990–2005.
Bugat R, Bataillard A, Lesimple T, Voigt JJ, Culine S, Lortholary A, et al. Summary of the Standards, Options and Recommendations for the management of patients with carcinoma of unknown primary site (2002). Br J Cancer. 2003;89(Suppl 1):S59-66.
Collado Martín R, GarcíaPalomo A, de la Cruz Merino L, BorregaGarcía P, Barón Duarte FJ. Spanish Society for Medical Oncology. Clinical guideline SEOM: cancer of unknown primary site. Clin Transl Oncol. 2014;16:1091–7.
Ettinger DS, Handorf CR, Agulnik M, Bowles DW, Cates JM, Cristea M, et al. Occult primary, version 3.2014. J Natl Compr Canc Netw. 2014;12:969–74 version 3.2014.
Fizazi K, Greco FA, Pavlidis N, Daugaard G, Oien K, Pentheroudakis G, et al. Cancers of unknown primary site: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v133–8.
NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Occult primary (cancer of unknown primary [CUP]). Version 1. 2022. National Comprehensive Cancer Network. 2021. https://www.nccn.org/professionals/physician_gls/pdf/occult.pdf . Accessed 12 Apr 2022.
Derakhshan S, Poosti A, Razavi AE, Moosavi MA, Mahdavi N, Naieni FB, et al. Evaluation of squamous cell carcinoma antigen 1 expression in oral squamous cell carcinoma (tumor cells and peritumoral T-lymphocytes) and verrucous carcinoma and comparison with normal oral mucosa. J Appl Oral Sci. 2021;29:e20210374.
Dall’Olio FG, Abbati F, Facchinetti F, Massucci M, Melotti B, Squadrilli A, et al. CEA and CYFRA 21–1 as prognostic biomarker and as a tool for treatment monitoring in advanced NSCLC treated with immune checkpoint inhibitors. Ther Adv Med Oncol. 2020;12:1758835920952994.
Campos-da-Paz M, Dórea JG, Galdino AS, Lacava ZGM, de Fatima Menezes Almeida Santos M. Carcinoembryonic antigen (CEA) and hepatic metastasis in colorectal cancer: update on biomarker for clinical and biotechnological approaches. Recent Pat Biotechnol. 2018;12:269–79.
Fujita T, Murayama K, Hanamura T, Okada T, Ito T, Harada M, et al. CSLEX (Sialyl Lewis X) is a useful tumor marker for monitoring of breast cancer patients. Jpn J Clin Oncol. 2011;41:394–9.
Kečkéš Š, Palaj J, Waczulíková I, Dyttert D, Mojtová E, Kováč G, et al. Pretreatment levels of chromogranin A and neuron-specific enolase in patients with gastroenteropancreatic neuroendocrine neoplasia. In Vivo. 2021;35:2863–8.
Fang L, Huang Z, Lin Y, Fu J, Liang X, Liu F. Clinical application of pro-gastrin-releasing peptide. Clin Lab. 2018;64:1259–68.
CAS PubMed Google Scholar
Tarao K, Nozaki A, Komatsu H, Komatsu T, Taguri M, Tanaka K, et al. Real impact of tumor marker AFP and PIVKA-II in detecting very small hepatocellular carcinoma (≤ 2 cm, Barcelona stage 0) - assessment with large number of cases. World J Hepatol. 2020;12:1046–54.
Umino K, Fujiwara SI, Ikeda T, Kawaguchi SI, Toda Y, Ito S, et al. Predictive value of soluble interlukin-2 receptor level at diagnosis on the outcome for patients with classical Hodgkin lymphoma treated with ABVD with or without radiotherapy. Ann Hematol. 2019;98:2121–9.
Lee T, Teng TZJ, Shelat VG. Carbohydrate antigen 19–9 - tumor marker: Past, present, and future. World J Gastrointest Surg. 2020;12:468–90.
Wang W, Xu X, Tian B, Wang Y, Du L, Sun T, et al. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta. 2017;470:51–5.
Sakurai Y, Kodaira S, Teramoto T, Sugano K, Abe O, Ohtake H, et al. Clinical evaluation of serum level of NCC-ST-439 as a new tumor marker for colorectal diseases–fluctuation of serum NCC-ST-439 in patients with colorectal cancers before and after surgery. Gan To Kagaku Ryoho. 1989;16:3205–12.
Lim JH, Park JS, Yoon DS. Preoperative fecal elastase-1 is a useful prognostic marker following curative resection of pancreatic cancer. HPB (Oxford). 2017;19:388–95.
Article Google Scholar
Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26–33.
Pentheroudakis G, Stoyianni A, Pavlidis N. Cancer of unknown primary patients with midline nodal distribution: midway between poor and favourable prognosis? Cancer Treat Rev. 2011;37:120–6.
Kodaira M, Yonemori K, Shimoi T, Yoshida A, Yoshida M, Kitano A, et al. Prognostic impact of presumed breast or ovarian cancer among patients with unfavorable-subset cancer of unknown primary site. BMC Cancer. 2018;18:176.
Download references
Acknowledgements
We thank all the patients whose data were used for the study. We thank Kyoko Onozawa for the secretarial assistance she provided. Editage (Cactus Communications) provided editorial support in the form of medical writing, table assembly, collating author comments, copyediting, fact-checking, referencing, and high-resolution image creation based on the authors’ detailed directions.
Author information
Authors and affiliations.
Department of Medical Oncology, National Cancer Center Hospital, 5-1-1 Tsukiji, Chuo-Ku, Tokyo, 104-0045, Japan
Shigemasa Takamizawa, Tatsunori Shimoi, Momoko Tokura, Shu Yazaki, Chiharu Mizoguchi, Ayumi Saito, Shosuke Kita, Kasumi Yamamoto, Yuki Kojima, Hitomi Sumiyoshi-Okuma, Tadaaki Nishikawa, Emi Noguchi, Kazuki Sudo & Kan Yonemori
Department of Diagnostic Pathology, National Cancer Center Hospital, 5-1-1 Tsukiji, Chuo-Ku, Tokyo, 104-0045, Japan
Masayuki Yoshida
You can also search for this author in PubMed Google Scholar
Contributions
All authors contributed to the study conception and design. Data collection and analysis were performed by ST. The first draft of the manuscript was written by ST and TS. All authors commented on previous versions of the manuscript. ST, TS, MY, MT, SY, CM, AS, SK, KY, YK, HO, TN, EN, KS, and KY read and approved the final manuscript.
Corresponding author
Correspondence to Tatsunori Shimoi .
Ethics declarations
Ethics approval and consent to participate.
The retrospective study protocol was approved by the National Cancer Center Research Ethics Review Committee (Tokyo, Japan; NCCH 2014–092), which waived the requirement for informed consent. All research was conducted in accordance with the Declaration of Helsinki. Patients are able to opt-out of the use of their data for research on the hospital’s website.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Takamizawa, S., Shimoi, T., Yoshida, M. et al. Diagnostic value of tumor markers in identifying favorable or unfavorable subsets in patients with cancer of unknown primary: a retrospective study. BMC Cancer 22 , 412 (2022). https://doi.org/10.1186/s12885-022-09514-3
Download citation
Received : 30 January 2022
Accepted : 07 April 2022
Published : 14 April 2022
DOI : https://doi.org/10.1186/s12885-022-09514-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Favorable subsets
- NCC-ST-439 (ST439)
- Sialyl-Tn (STN)
- Tumor marker
- Unfavorable subsets
ISSN: 1471-2407
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Tumor Markers
What are tumor markers.
A tumor marker is anything present in or produced by cancer cells or other cells of the body in response to cancer or certain benign (noncancerous) conditions that provides information about a cancer, such as how aggressive it is, what kind of treatment it may respond to, or whether it is responding to treatment.
Tumor markers have traditionally been proteins or other substances that are made at higher amounts by cancer cells than normal cells. These can be found in the blood, urine, stool , tumors, or other tissues or bodily fluids of some patients with cancer.
Increasingly, however, genomic markers (such as tumor gene mutations, patterns of tumor gene expression , and nongenetic changes in tumor DNA) are being used as tumor markers. These markers are found both in tumors themselves and in tumor fragments shed into bodily fluids.
Many different tumor markers have been characterized and are in clinical use . Some are associated with only one type of cancer, whereas others are associated with multiple cancer types.
How are tumor markers used in cancer care?
Tumor markers can provide a wide variety of information that is important for cancer care, such as
- Helping to diagnose cancer. However, having an elevated level of a tumor marker does not mean that someone has cancer. Noncancerous conditions can sometimes cause an increase in the level of a tumor marker. In addition, not everyone with a particular type of cancer will have a higher level of a tumor marker associated with that cancer. Therefore, measurements of tumor markers are usually combined with the results of other tests, such as biopsies or imaging, to diagnose cancer.
- The type of cancer
- The stage of the cancer
- An estimate of prognosis
- What treatment may be effective. Tumor markers that indicate whether someone is a candidate for a particular targeted therapy are sometimes referred to as biomarkers for cancer treatment . Biomarkers are generally measured in samples of tumor tissue. However, tumors can shed cells or bits of biological material into blood, and these can be measured by tests called liquid biopsies .
- How well the treatment is working. Periodic (or “serial”) measurements of a marker made while someone is undergoing treatment can indicate whether the tumor is responding to treatment.
- Whether cancer has returned. Measuring tumor markers periodically after treatment has ended may be used to check for recurrence.

Have Questions?
Connect with our cancer information specialists.
Phone: 1-800-4-CANCER Chat: LiveHelp Email: [email protected]
Available Monday–Friday 9:00 a.m. to 9:00 p.m. ET.
What tumor marker tests are currently being used, and for which cancer types?
A number of tumor marker tests are currently being used for a wide range of cancer types. See the list of tumor marker tests in common use for more information.
Many tumor marker tests are carried out by commercial and academic laboratories. Sometimes cancer centers use a tumor marker test developed within a single clinical laboratory (also known as a lab-developed test or LDT) to meet a specific medical need. All tumor markers, including those tested by LDTs , are tested in laboratories that meet standards set by the Clinical Laboratory Improvement Amendments program .
Can tumor markers be used to detect cancer in people who don't have symptoms?
Because tumors produce markers that can be measured in blood and other body fluids, researchers have hoped that they might also be useful in screening people for cancer—that is, detecting cancer at an early stage before it causes symptoms.
However, studies to see whether circulating tumor markers can be used to screen for cancer have generally found that these markers do not work well for screening. They often don’t identify everyone with the disease (they are not sensitive enough). Or they may indicate the possible presence of cancer in people who don’t actually have cancer (they are not specific enough).
Researchers are now testing whether multi-cancer detection tests (MCDs), which analyze multiple biomarkers in the blood of people without symptoms, can identify early cancers.
Most MCDs examine DNA that tumor cells release into blood. They may also analyze other biological molecules in blood, such as proteins. Tests that look at tumor markers in blood and other body fluids are sometimes called liquid biopsy tests.
Although many MCD tests are in development and several are already being marketed, much remains to be learned about how best to use these tests and their harms and benefits. A critical question is whether treating the cancers identified by MCD tests would reduce deaths from these cancers. NCI will be launching a clinical trial to better understand the implications of using MCD tests to screen for cancer .
What research is under way to develop additional tumor markers?
NCI’s Early Detection Research Network (EDRN), a collaborative consortium of academic and private-sector investigators, has focused on the systematic discovery, development, and validation of biomarkers and imaging methods to detect early-stage cancers and to assess risk for developing cancer. One goal of EDRN is to develop biomarkers that can distinguish aggressive early-stage cancers from slow-growing cancers that would never cause symptoms to reduce overtreatment .
Cancer researchers are turning to proteomics (the study of protein structure, function, and patterns of expression) and proteogenomics (the integration of proteomics with genomics and gene expression analysis, or transcriptomics ) with the hope of developing novel biomarkers that can be used to identify cancer in its early stages, to predict the effectiveness of treatment, and to predict the chance of cancer recurrence.
Artificial intelligence and machine learning are increasingly being investigated, including by EDRN, as tools to analyze and interpret patterns in genomic and proteomic markers that help predict cancer risk and diagnose specific cancer types.
NCI’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) is using a proteogenomic approach for tumor marker discovery for a growing number of cancers, including colorectal, breast, and ovarian cancers. By systematically identifying proteins (and associated biological processes) that originate from alterations in cancer genomes, CPTAC researchers have discovered new tumor subtypes, tumor microenvironment variations, and new potential proteins for targeted drug therapy. Recent innovations have suggested that these analyses could be done on a microscale using very small amounts of tumor tissue obtain from a biopsy.
The NCI Cancer Moonshot SM Biobank is working with patient participants at community hospitals around the country to encourage them to donate tissue and blood samples over the course of their cancer treatment. The samples are sent to researchers who use them to better understand cancer and potentially identify tumor markers.
More information on NCI’s role in supporting research on novel tools and methods for diagnosing cancer is available on the Cancer Diagnosis Research page.
An official website of the United States government
Here’s how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you’ve safely connected to the .gov website. Share sensitive information only on official, secure websites.

- Health Topics
- Drugs & Supplements
- Medical Tests
- Medical Encyclopedia
- About MedlinePlus
- Customer Support
Tumor Marker Tests
What are tumor marker tests.
These tests look for tumor markers, which are sometimes called cancer markers. Tumor markers are substances that are often made by cancer cells or by normal cells in response to cancer . For example, some tumor markers are proteins that certain cancer cells make in larger amounts than normal cells do. Changes in the genes and other parts of tumor cells can be tumor markers, too.
Certain tumor markers may be found in samples of body fluids, such as blood or urine (pee). Other tumor markers are found in samples of cells that are removed from a tumor during a biopsy .
Tumor marker tests are mainly used after you have a cancer diagnosis. The test results may help answer important questions about your cancer, such as:
- How fast is the cancer growing?
- What type of treatment is most likely to help?
- Is the treatment really working?
- Has cancer come back after treatment?
Not all cancers have known tumor markers. And the tumor markers that are known don't provide perfect information. That's because:
- Some conditions that aren't cancer may also cause high levels of certain tumor markers. Tumor marker tests can't tell whether tumor markers come from cancer or from another condition.
- Some people don't make high levels of the tumor markers that are commonly found in their type of cancer.
But even with these limits, tumor marker testing can often give a more complete picture of your cancer when they are used along with the results of other tests and exams.
What are they used for?
Tumor marker tests are mainly used to learn more about a known cancer. But in certain cases, they may be used to screen for cancer or to help diagnose the disease.
Tumor marker tests are most often used after you have a cancer diagnosis. When used with other tests, tumor markers may help:
- Find out whether cancer has spread to other parts of your body (cancer stage)
- Predict how fast your cancer may grow, the chance of recovery, and whether cancer is likely to return
- Select the right treatment for your type of cancer. Some treatments work only with cancers that have certain tumor markers. Tumor markers that help plan treatment are also called biomarkers.
- Monitor how well your treatment is working. If tumor marker levels go down, it usually means your treatment is helping.
- Find any cancer that remains after treatment or cancer that comes back after treatment.
Some tumor marker tests that use body fluids (mainly blood or urine) have a limited role in screening for certain types of cancer. The tests are mostly used to screen people who:
- Have a high risk for the type of cancer that's linked to the tumor marker being measured
- Have symptoms that could be from that type of cancer.
Tumor marker tests that are used to screen for cancer can't diagnose cancer. If you have a high level of tumor markers, it only means that you're more likely to have cancer. A biopsy is usually needed to diagnose or rule out cancer.
Tumor marker tests that use cells from a tumor may help diagnose cancer. These "tumor cell markers" are usually removed during a biopsy. They may be used with other tests to confirm a cancer diagnosis and decide on the best treatment.
Why do I need a tumor marker test?
You may need a tumor marker test if you:
- Are currently being treated for cancer
- Have finished cancer treatment
- Have a high risk of getting a certain type of cancer because it runs in your family, or you have other conditions that increase your risk
The type of test you have will depend on your health and health history, cancer diagnosis, and the symptoms you may have. Below are some of the most common types of tumor markers and how they are used. Some tumor markers are linked with only one type of cancer. Others are linked with many different types of cancers:
What happens during a tumor marker test?
Blood tests are the most common type of tumor marker tests. Urine tests or biopsies are also frequently used to check for tumor markers. A biopsy is a minor procedure that involves removing a small piece of tissue for testing.
For a blood test: A health care professional will take a blood sample from a vein in your arm, using a small needle. After the needle is inserted, a small amount of blood will be collected into a test tube or vial. You may feel a little sting when the needle goes in or out. This usually takes less than five minutes.
For a urine test: Your health care provider will tell you how to provide your sample.
For a biopsy: A provider will remove a small piece of tissue. There are many ways to do a biopsy, depending on where the sample is located. A biopsy of your skin may be done by cutting or scraping the area. A biopsy of tissue from inside your body may use a special needle to remove a sample or a small incision (cut) to remove all or part of a suspicious area.
Will I need to do anything to prepare for the test?
For a blood or urine test: You usually don't need any special preparations.
For a biopsy: You may need to fast (not eat or drink) for several hours before the procedure.
Talk with your provider if you have any questions about preparing for your test.
Are there any risks to the test?
For a blood test: There is very little risk to having a blood test. You may have slight pain or bruising at the spot where the needle was put in, but most symptoms go away quickly.
For a urine test: There is no risk to a urine test.
For a biopsy: You may have a little bruising or bleeding at biopsy site. You may also have a little discomfort for a day or two.
What do the results mean?
Your provider will review your tumor marker test results along with other information about your condition. Together, you can discuss how your results affect your diagnosis, treatment, and schedule for future testing.
Learn more about laboratory tests, reference ranges, and understanding results .
- Cancer.Net [Internet]. Alexandra (VA): American Society of Clinical Oncology; 2005-2022. Tumor Marker Tests; [updated 2020 May; cited 2022 Jun 16]; [about 4 screens]. Available from: https://www.cancer.net/navigating-cancer-care/diagnosing-cancer/tests-and-procedures/tumor-marker-tests
- Hinkle J, Cheever K. Brunner & Suddarth's Handbook of Laboratory and Diagnostic Tests. 2nd Ed, Kindle. Philadelphia: Wolters Kluwer Health, Lippincott Williams & Wilkins; c2014. Cancer Tumor Markers (CA 15-3 [27, 29], CA 19-9, CA-125, and CA-50); 121 p.
- Merck Manual Consumer Version [Internet]. Kenilworth (NJ): Merck & Co. Inc.; c2022. Diagnosis of Cancer; [updated 2020 Sep; cited 2022 Jun 16]; [about 4 screens]. Available from: https://www.merckmanuals.com/home/cancer/overview-of-cancer/diagnosis-of-cancer
- National Cancer Institute [Internet]. Bethesda (MD): U.S. Department of Health and Human Services; Tumor Markers; [updated 2021 May 11; cited 2022 Jun 16]; [about 4 screens]. Available from: https://www.cancer.gov/about-cancer/diagnosis-staging/diagnosis/tumor-markers-fact-sheet#q1
- National Heart, Lung, and Blood Institute [Internet]. Bethesda (MD): U.S. Department of Health and Human Services; Blood Tests; [updated 2022 Mar 24; cited 2022 Jun 16]; [about 7 screens]. Available from: https://www.nhlbi.nih.gov/health/blood-tests
- Oncolink [Internet]. Philadelphia: Trustees of the University of Pennsylvania; c2022. Patient Guide to Tumor Markers; [updated 2022 Mar 1; cited 2022 Jun 16]; [about 3 screens]. Available from: https://www.oncolink.org/cancer-treatment/procedures-diagnostic-tests/blood-tests-tumor-diagnostic-tests/patient-guide-to-tumor-markers
- Testing.com [Internet]. Seattle (WA): OneCare Media; c2022. Biopsy; [cited 2022 Jun 16]; [about 1 screen]. Available from: https://www.testing.com/glossary/#biopsy
- Testing.com [Internet]. Seattle (WA): OneCare Media; c2022. Tumor Markers; [updated 2021 Nov 9; cited 2022 Jun 16]; [about 12 screens]. Available from: https://www.testing.com/tests/tumor-markers/
- University of Rochester Medical Center [Internet]. Rochester (NY): University of Rochester Medical Center; c2022. Health Encyclopedia: Lab Tests for Cancer; [cited 2022 Jun 16]; [about 6 screens]. Available from: https://www.urmc.rochester.edu/encyclopedia/content.aspx?contenttypeid=85&contentid=p07248
- UW Health: American Family Children's Hospital [Internet]. Madison (WI): University of Wisconsin Hospitals and Clinics Authority; c2022. Kids Health: Biopsies; [updated 2019 Jul 1; cited 2022 Jun 16]; [about 3 screens]. Available from: https://patient.uwhealth.org/kidshealth/en/parents/biopsy.html/article
- UW Health [Internet]. Madison (WI): University of Wisconsin Hospitals and Clinics Authority; c2022. Tumor Markers: Topic Overview; [updated 2021 Sep 8; cited 2022 Jun 16]; [about 3 screens]. Available from: https://patient.uwhealth.org/healthwise/article/en-us/abq3994
The information on this site should not be used as a substitute for professional medical care or advice. Contact a health care provider if you have questions about your health.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- World J Gastroenterol
- v.22(5); 2016 Feb 7
Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances
Correspondence to: Gustaw Lech, MD, PhD, Department of General, Gastroenterological and Oncological Surgery, Medical University of Warsaw, 1a Banacha Street, 02097 Warsaw, Poland. [email protected]
Telephone: +48-22-5992482 Fax: +48-22-5992057
Colorectal cancer (CRC) is the second most commonly diagnosed cancer among females and third among males worldwide. It also contributes significantly to cancer-related deaths, despite the continuous progress in diagnostic and therapeutic methods. Biomarkers currently play an important role in the detection and treatment of patients with colorectal cancer. Risk stratification for screening might be augmented by finding new biomarkers which alone or as a complement of existing tests might recognize either the predisposition or early stage of the disease. Biomarkers have also the potential to change diagnostic and treatment algorithms by selecting the proper chemotherapeutic drugs across a broad spectrum of patients. There are attempts to personalise chemotherapy based on presence or absence of specific biomarkers. In this review, we update review published last year and describe our understanding of tumour markers and biomarkers role in CRC screening, diagnosis, treatment and follow-up. Goal of future research is to identify those biomarkers that could allow a non-invasive and cost-effective diagnosis, as well as to recognise the best prognostic panel and define the predictive biomarkers for available treatments.
Core tip: This review summarizes data concerning clinical utility of biomarkers in colorectal cancer patients. Authors focus primarily on currently available diagnostic, prognostic and predictive biomarkers of the disease. Great attention is also paid to the advances achieved in personalized therapy of colorectal cancer.
INTRODUCTION
Worldwide, colorectal cancer (CRC) annually affects more than one million men and women and causes more than half a million deaths[ 1 ]. In Europe in 2010, CRC was the third most common malignant cancer in both men and women[ 2 ]. There are 250000 cases of colorectal cancer diagnosed on an annual basis in Europe only. Five-year survival was 54 percent among adult Europeans diagnosed with colorectal cancer between 1995 and 1999[ 3 ]. More recent available data report that the overall five-year relative survival can achieve 65 percent, but varies depending on stage of cancer disease[ 4 ].
The number of biomarkers used for tests continues to grow. The National Institute of Health defines a biomarker as a biological molecule found in blood, other body fluids, or tissues that is a sign of normal or abnormal process, or of a condition or disease[ 5 ]. A definition of biomarker mostly refers to DNA, RNA, microRNA (miRNA), epigenetic changes or antibodies. A term tumour marker, by some researchers considered as a synonym of biomarker, refers to substances (most typically proteins, glycolipids) representing biological structures, which can be attributed to the development of normal cells or carcinogenesis at different cell development stages e.g ., tumour-associated antigens (TAAs) which are the largest group of clinically significant markers. As a result, the concentration of TAAs typically correlates with the number (or mass) of specific neoplastic cells.
In daily clinical practice, in the process of diagnosis and therapy, there are several parameters in use of long-established high sensitivity, specificity and positive predictive value. These parameters have been selected from among tens of molecules produced by cells in long-term laboratory tests, observational studies and clinical trials. The concentrations of tumour markers tested at the diagnostic stage are believed to assist in early cancer diagnosis and to be used in screening tests. Some of them are currently found to be more important during treatment and long-term follow-up. On the other hand, for some types of tumours, markers are also considered important in monitoring the progress of treatment, efficacy of neo-adjuvant therapy, surgery, adjuvant chemotherapy and radiation therapy and follow-up for possible recurrence. Long-term observational studies also point to the fact that, apart from determining antigen concentration, it can be also important to trace its progress and dynamics. In this review, we have updated a review published in 2014[ 6 ]. We examine molecular (genetic, epigenetic, protein) biomarkers associated with CRC and discuss their role in cancer screening, early detecting of disease recurrence and as prognostic and predictive factors.
BLOOD AND STOOL MARKERS FOR COLORECTAL CANCER SCREENING AND FOLLOW UP
Blood and stool genetic and epigenetic markers.
Several authors have investigated molecular non-invasive screening tests for early detection of CRC. DNA, RNA and other molecules derived by tumour in stool, as well as their concentrations in blood have been studied extensively. Colorectal process of carcinogenesis is characterized by genetic and epigenetic alteration transforming normal cells into cancer cells. Most studies concerning molecular markers in stool have focused on the detection of tumour DNA. These investigations have concentrated on the detection of mutated KRAS , TP53 , APC and markers for microsatellite instability (MSI)[ 7 - 9 ]. A faecal DNA test targeted at molecular biomarkers has been commercially available for twelve years, with reported sensitivity for cancer ranging from 25% up to 92% for the latest tests based on BEAMing technology, and 94%-98% specificity[ 10 - 12 ]. Apart from genetic alterations, the DNA promoter hypermethylation silencing the tumour suppressor genes has been widely investigated. Epigenetic changes, depending on the markers or their combinations evaluated, have been detected in CRC patients with 70%-96% sensitivity and 72%-96% specificity[ 9 , 13 , 14 ]. Many combinations of genetic and epigenetic markers have been studied, but until now, the results have not endorsed their use in clinical practice. Using blood instead of stool as a screening material could offer some obvious advantages. Several studies have evaluated potential plasma DNA genetic and epigenetic biomarkers in CRC detection. The overall sensitivity ranges from 30% to 87%, with specificity of up to 96%. The use of RNA biomarkers in stool has not been investigated as extensively as was the case for DNA biomarkers, mainly because stool environment is responsible for mRNA degradation, although improving laboratory retrieval methods seems to solve this problem. Koga et al[ 15 ] analysed mRNA expression of MMP7 , PTGS2 , TP53 and MYBL2 in colonocytes isolated from stool by quantitative real-time RT-PCR, to find out that these markers can identify CRC patients with 58% sensitivity and 88% specificity. Sensitivity was found to depend on tumour size and tumour location, but not cancer stage[ 15 ]. Most recently, the so called transcriptomic studies have investigated the expression of miRNAs - short, non-coding 18-22 nucleotide RNA molecules in stools of CRC patients. The most extensively studied miR21, miR106a, miR135, miR17-92 were found to be overexpressed in CRC patients compared with healthy individuals[ 16 , 17 ]. As was the case with RNA markers in stool, many studies have been evaluating mRNA of different tumour genes in whole blood, plasma or circulating tumour cells to identify new CRC screening markers. Most of them investigated mRNA molecules of CK19, CK20, or Carcinoembryonic antigen (CEA). The overall sensitivity of these markers was up to 72%, specifically when combinations of these markers were used[ 18 , 19 ]. The specificity was very high with healthy control samples or much lower when compared to other cancer or inflammatory bowel diseases samples[ 20 ]. Recent studies have indicated that circulating miRNAs may be involved in the process of oncogenesis. The use of miRNA as a biomarker is now being evaluated. A large number of miRNA molecules have been assessed, with a focus on miR145, miR143, miR135, miR17-92. More specifically, Huang et al[ 21 ] has found that plasma miR29a and miR92a demonstrated a significant diagnostic value for advanced neoplasia with 83% and 84% sensitivity and specificity, respectively, in discriminating CRC patients. These studies need to be validated in randomised trials to define their value in CRC screening.
Blood and stool protein markers
Protein markers for screening and early detection of CRC can be divided into tumour TAAs, antibodies against TAAs, and other CRC-relevant proteins. CEA was discovered almost 50 years ago, in 1965, and it still remains the only tumour marker of recognised efficacy in monitoring CRC patients’ therapy[ 22 ]. CEA was first considered specific for CRC, but elevated CEA levels were later detected in other neoplasms too, e.g . gastric and pancreatic cancers, and in inflammatory conditions. Elevated CEA concentrations are only rarely identified in CRC stage I. Moreover, CEA does not differentiate benign versus malignant polyps. According to The European Group on Tumor Markers, European Society of Medical Oncology and American Society of Clinical Oncology guidelines[ 2 , 23 , 24 ], CEA is not recommended for use in screening tests. Recently, some studies have investigated the advantages of mRNA molecules encoding CEA for the detection of CRC, but the results were not superior to those of CEA[ 19 ].
In some studies, high CEA concentrations in patients with CRC stage II and III were found to be potentially indicative of more aggressive types of cancer[ 25 , 26 ]. Earlier, the Colorectal Working Group of American Joint Committee on Cancer proposed to include CEA baseline concentration to the traditional TNM classification as the so-called C-stage. C-stage was proposed to be divided into Cx, C0 (CEA < 5 ng/mL) and C1 (CEA > 5 ng/mL) substages[ 27 ]. The meaning of CEA as an independent prognostic factor was also confirmed in a recent retrospective analysis of 17910 patients with CRC, with a mean 27-mo follow-up, with longer survival periods for patients with IIA C0 and IIIA C0 vs I C1, IIIA C0 vs IIA C1, and IIIB C0 vs IIB-C C1, respectively[ 28 ]. No study, however, has shown that CEA concentration level can be used to select those patients with stage II CRC who would benefit from adjuvant chemotherapy. From a prognostic point of view, it appears reasonable to determine CEA levels before surgery in patients with disseminated CRC. The roles of CEA in determining life expectancy was confirmed in several studies on patients with liver metastases[ 29 , 30 ]. Recent study proved that combined use of CEA and serum amyloid A (SAA) is able to identify patients with favourable and poor prognosis. In addition to tumour baseline parameters, routine analysis of CEA together with SAA provides improved prognosis value on cancer specific survival and disease-free survival in resected rectal cancers[ 31 ]. CEA half-life is known to last approximately 7 d. After R0 resection surgery, CEA levels should return to normal within 4 to 6 wk. Sustained elevated CEA levels can be indicative of infiltration or metastases. Slow increase in CEA concentrations after surgery is a typical sign of local recurrence, whereas dynamically increasing levels can be symptomatic of metastases, most probably in the liver[ 24 , 32 ]. Testing CEA levels is considered most cost-effective in detecting post-surgery recurrences[ 24 ]. Please note that CEA levels tested every 3 mo for the first 3 years and thereafter every 6 mo for subsequent 2-3 years is a golden follow-up standard after CRC therapy recommended by a number of scientific associations[ 2 , 23 , 33 ]. It appears particularly important in asymptomatic patients, in whom chemotherapy can be used, with a much longer life expectancy as compared to treatment administered after the onset of symptoms of recurrence. CEA is a marker of choice in monitoring disseminated disease during systemic therapy. Constant increase in CEA levels is typically associated with a progression of the disease, even though radiological tests may prove otherwise[ 23 , 24 ]. However, chemotherapy can also result in temporary increase in CEA concentration, which must be also taken into account. Therefore, it is not recommended to test CEA levels within 2 wk of chemotherapy, whereas in patients on oxaliplatin, tests can be carried out after 4 to 6 wk.
Cancer antigen 19-9 (CA 19-9) is a glycoprotein whose relevance in CRC diagnosis still remains an issue. The majority of researchers arrived at the conclusion that CA 19-9 sensitivity is much inferior to that of CEA, and that elevated CA 19-9 levels is a poor prognostic factor[ 2 , 23 , 34 - 36 ]. Other carbohydrate antigens: CA 195, CA 50 have been also investigated, but with comparatively disappointing results. CA 72-4 is a biomarker with poor sensitivity ranging from 9% to 31% and better specificity ranging from 89% to 95% in patients screened for CRC. The diagnostic information in recurrent CRC provided by CA 72-4 has borderline significance, by far worse than CEA. All authors conclude that CA 72-4 sensitivity is rather low and specificity incomplete in screening and following up in patients with CRC. On the other hand, an algorithm based on combination of CEA, CA 19-9, CA 72-4, CA 242, CYFRA21-1 improves the diagnostic accuracy compared with these biomarkers alone[ 34 - 39 ]. Among other protein markers examined for screening purposes, two have been extensively investigated: the tumour specific M2 isoform of pyruvate kinase (M2-PK) in stool and tissue inhibitor of matrix metalloproteinase 1 (TIMP1). M2-PK measured in stool showed relatively high sensitivity for CRC up to 91%, and much lower for adenomas[ 40 , 41 ]. Plasma level of TIMP1 is reported to be elevated in CRC patients and prospective studies have been carried to assess its utility as biomarker. The results of the study included more than 4500 patients screened by endoscopy for CRC demonstrated that TIMP1 is not significantly superior to CEA marker in cancer screening and is not suitable for the detection of premalignant lesions[ 42 ]. Tissue polypeptide-specific antigen (TPS) and tissue polypeptide antigen (TPA) which detects the fragments of cytokeratines 8, 18 and 19 due to lack of sensitivity and specificity can not to be recommended in CRC screening. The majority of investigators have found that increased levels of TPA and TPS are observed in metastatic stage of CRC. A further studies has suggested that combination of TPA and CEA rises the sensitivity of these biomarkers in identifying the patients with CRC recurrence[ 34 , 37 , 43 , 44 ]. Other biomarkers, such as: thymidine phosphorylase (TP), DNA ploidy were determined to be insignificant in detecting, staging and following-up of patients with CRC[ 23 ].
MOLECULAR PROGNOSTIC AND PREDICTIVE BIOMARKERS
With the recent progress in understanding the molecular mechanisms of cancer development, dissemination, resistance to chemotherapy, and radiation therapy, it is now easier to select the most proper strategy for managing CRC. Clinical prospective and retrospective studies open the door for biomarkers use in clinical practice to assist in selecting the best drugs, both standard, such as 5-fluorouracil, oxaliplatin or irinotecan, and new generation targeted drugs: cetuximab, panitumumab, or bevacizumab. Biomarker identification is particularly important for patients with CRC stage II. In this group of patients, the risk of recurrence is only 20 percent. It is also desirable to use adjuvant therapy in this type of patients. There are attempts to select this group of patients based on genetic tests, or to personalise chemotherapy based on specific biomarkers. The following markers discovered throughout the recent years continue to be closely examined: MSI, chromosome 18q loss of heterozygosity (18qLOH), p53 , KRAS , BRAF , NRAS , PIK3CA mutations, PTEN expression, UGT1A1 gene polymorphism, and ezrin protein (Table (Table1 1 ).
Recommendations for use of tumour markers and biomarkers in colorectal cancer by groups of experts
CEA: Carcinoembryonic antigen; MSI: Microsatellite instability; 18qLOH: Chromosome 18q loss of heterozygosity; VEGF: Vascular endothelial growth factor; TPS: Tissue polypeptide-specific antigen; TPA: Tissue polypeptide antigen.
MSI denotes changes in coding and non-coding sequences of microsatellite chromosomes, i.e . repeated DNA sequences. These sequences are particularly exposed to errors in the mutation repair system that consist in the loss or multiplication of nucleotide sequence repetitions, which results in shortening or extension of microsatellite regions in neoplastic cells. Mutations arising out of these processes are eliminated by mismatch repair genes ( MMR ) such as MSH2 , MSH6 , PMS2 and MLH1 , which makes some researchers believe that MSI can be caused by mutations in these genes[ 45 ]. Microsatellite instability can be classified into microsatellite instability-high (MSI-H), and microsatellite instability-low (MSI-L), depending on the percentage of loci that correlate to MSI characteristics. Tumour cells that lack MSI features are designated as MSS.
In retrospective studies and meta-analyses in patients with CRC stage II and III, MSI-H was shown to be a predictive factor that improved overall survival (OS), irrespective of the progression (stage) of cancer. A lower incidence of lymph node metastases and distant metastases as compared to MSI-L or MSS cancer cells was also observed[ 46 - 49 ]. MSI status is currently recommended in the WHO classification of mucinous-type CRC - MSI-H indicates good prognosis, MSI-L or MSS - poor outcome. However, MSI should be considered more of a prognostic rather than predictive factor. This conclusion is based on equivocal results of studies evaluating the efficacy of 5-FU-based chemotherapy in groups of patients with MSI-H and MSI-L or MSS. Ribic et al[ 48 ] examined tumour specimens collected from 570 patients with CRC stage II and III and correlated the test results with chemotherapy outcomes in these patients to reveal a tendency for shorter overall survival in patients with MSI-H on adjuvant therapy. Significant improvement was observed in patients with MSS tumours. A recent pooled analysis of randomized clinical studies revealed significant decrease in the overall five-year survival rate for patients with CRC stage II and MSI-H on 5-FU-based chemotherapy. 5-FU-based chemotherapy was found to improve therapeutic outcomes only in patients with CRC stage III and MSI-L or MSS[ 50 ]. Some studies indicated potentially negative effects of 5-FU-based chemotherapy in patients with MSI-H. A longer survival rate as compared to patients on 5-FU-based adjuvant chemotherapy was observed in a reference group of patients undergoing surgical treatment. Resistance of MSI-H tumours to 5-FU was also confirmed in in vitro studies[ 51 ]. A completely different conclusion can be drawn from earlier studies of Elsaleh et al[ 52 , 53 ], which confirmed the efficacy of 5-FU in patients with CRC stage III and MSI-H. Recent study also proved that prognostic value of MMR mutation was similar in the presence or absence of fluorouracil and folinic acid chemotherapy[ 54 ]. Beragnolli et al[ 55 ] revealed that a higher rate of overall 5-year progression-free survival was observed in patients with CRC stage III and MSI-H on 5-FU and irinotecan vs 5-FU-based chemotherapy. To recap, the results of MSI studies and clinical experience in patients with CRC stage II indicate that the degree of microsatellite instability may be of significance as a prognostic factor. Also, adjuvant 5-FU-based chemotherapy was proved to provide no benefits (or potentially cause adverse reactions) in patients with MSI-H. Further research is needed to investigate whether the MSI status can predict benefit (in high-risk patients) from irinotecan-based treatment or oxaliplatinum-based therapy.
Chromosome 18q loss of heterozygosity
A number of studies were dedicated to another prognostic factor in patients with CRC stage II and III - chromosome 18q loss of heterozygosity in the coding place of, inter alia, SMAD 4 proteins specific to CRC. In these studies, the overall 5-year survival was poorer for patients with CRC stage III and 18qLOH as compared to non-18qLOH patients[ 56 ]. A meta-analysis of data from 27 studies and 2189 patients by Popat et al[ 57 ] confirmed that poorer survival was correlated with 18q chromosome deletion. Two years later, the same research team questioned these findings after re-examining the same data[ 58 ]. Likewise, no correlation was identified between the presence of 18qLOH and 5-year survival in patients with non-MSI-H phenotype[ 59 ]. The role of 18qLOH in predicting response to standard chemotherapy has not been yet fully confirmed. Watanabe et al[ 60 ] demonstrated better response to 5-FU-based chemotherapy in patients with CRC stage III and MSS and with the absence of 18q chromosome deletion vs. patients in whom 18q chromosome deletion was present. The recently published results of the same research team can be a proof that in patients with CRC stage II and III and MSS-H (> 33%), the level of LOH of four chromosomes, including 18, is correlated with significantly poorer survival rate as compared to patients with MSS and LOH-L or MSI-H phenotype[ 61 ].
Based on the available data, 18q chromosome deletion cannot be the sole basis for any therapeutic decisions, however, it is being more closely examined under ECOG 5202 study, featuring molecular markers identified so far in selecting the most proper adjuvant post-surgery treatment, by prospectively analysing the role of MSI and 18qLOH in prognosis and therapeutic decisions in patients with CRC stage II. Patients with good prognosis (with MSI-H and w/o 18qLOH) were followed-up, and patients with poor prognosis (with MSI-L or MSS and 18qLOH) were randomized to one of two groups on chemotherapy (FOLFOX alone or FOLFOX and bevacizumab). The results of E5202 are expected in the next few years. No conclusion can be drawn from this study about the possible inefficacy of chemotherapy in patients with MSI-H, however, the study will include a multifactor analysis of biomarkers that can assist in taking therapeutic decisions in other groups of patients[ 62 ].
P53 mutation
Mutation in the tumour suppressor gene p53 (chromosome region 17p13) occur in 50%-70% of all CRC and is associated with worse outcomes, including disease free survival and overall survival[ 63 ]. Results obtained from a study that included more than 3500 CRC patients confirm the prognostic value of p53 mutation, which seems to be determined by the primary tumour site. Patients with p53 mutation and tumour of proximal colon had better OS when treated with adjuvant chemotherapy compared to those treated by surgery alone[ 64 ].
Biomarkers suitable in anti-epidermal growth factor receptor therapy
A number of currently tested markers have been discovered in the course of studies on epidermal growth factor receptor (EGFR) signalling pathways. KRAS gene mutation on short arm of chromosome 12 at codon 12 (80% of patients) or, to a lesser extent, codon 13 is believed to be of use as a biomarker in patients on cetuximab or panitumumab[ 65 ]. These mutations are one of the most common in proliferative diseases (37% and 13%, respectively), and their significance in CRC carcinogenesis has been examined in much detail[ 66 ]. As these mutations are present in EGFR signalling pathway, they can be a predictive factor for therapy with anti-EGFR antibodies. In studies performed so far, KRAS mutation was found to be correlated with non-responsiveness to cetuximab and panitumumab[ 67 , 68 ]. CRYSTAL and OPUS data indicate that the effectiveness of FOLFOX or FOLFIRI alone is no inferior to that of cetuximab in patients with KRAS in combination with chemotherapy according to FOLFIRI and FOLFOX regimen, respectively. However, in non-KRAS patients, cetuximab improves the therapeutic outcome[ 69 , 70 ]. The same conclusions can be drawn from the results of other large clinical studies: COIN, NORDIC VII or PRIME[ 71 - 73 ]. Yet, the effects of KRAS mutation at codon 12 or 13 on tumour biology were found to differ. In two studies, the survival rate was higher in patients with an uncommon G13D mutation at codon 13 on cetuximab vs patients with other mutations, and similar to patients with no KRAS mutations identified[ 65 , 74 ]. It is presently believed that anti-EGFR antigens should not be used in patients with tumours indicative of G12V mutation of KRAS at codon 12. For bevacizumab, KRAS mutation was found to be of no use as a predictive factor[ 75 ].
The same applies to BRAF mutations found in 8%-13% of patients with CRC, which are mutually exclusive with KRAS mutations. The most frequently observed BRAF mutation is V600E mutation. BRAF mutations make the tumour to a large extent resistant to anti-EGFR monoclonal antibodies, and significantly worsen prognosis, especially in patients with MSI-L and MSS[ 66 , 70 , 76 - 78 ]. Based on the available data, National Comprehensive Cancer Network (NCCN) suggests considering BRAF mutation testing when KRAS is mutation negative. Interestingly, good prognosis was reported even in those MSI-H CRC patients who had coincident BRAF mutations[ 78 ]. In one of studies, the OS period was shown to be slightly longer in patients on cetuximab even if the BRAF mutation was present[ 69 ]. Very limited response to vemurafenib, recently approved for metastatic melanoma patients harboring BRAF (V660E) mutation, was demonstrated in CRC patients. Researchers reported that by adding cetuximab strongly synergistic reaction with BRAF inhibitors was observed[ 79 ]. NRAS is another member of RAS proto-oncogenes which was found to be rarely mutated, while BRAF is mutually exclusive with KRAS mutations. Since NRAS mutation can predict resistance to EGFR therapy, NCCN suggests considering NRAS mutation testing when KRAS is mutation negative. To date, NRAS mutation does not appear to be associated with the prognosis[ 80 ].
Phosphatidylinositide-3-kinases (PI3K) are kinases that promote cellular proliferation. Mutations in PIK3CA gene encoding p110α catalytic subunit of PI3K have been identified in different human solid tumours, including CRC. PIK3CA gene is mutated in 10%-20% of CRC tumours. PIK3CA gene encodes the kinase that regulates, alongside with KRAS, downstream signalling pathways of EGFR. Moreover, PI3K-initiated signalling is inhibited by phosphatase and tensin homologue deleted on chromosome 10 ( PTEN ). Recent studies have revealed an increase in colon cancer-specific mortality in patients with PIK3CA-mutated tumours, as compared with patients with PIK3CA wild-type tumours[ 81 , 82 ]. However only the coexistence of PIK3CA exon 9 and 20 mutations but not PIK3CA mutation in either exon 9 or 20 alone has been reported to be associated with the worse prognosis[ 82 ]. Among patients with KRAS wild-type tumours, the presence of PIK3CA mutation correlated with a significant increase in CRC specific mortality. In contrast, PIK3CA mutation did not significantly affect mortality among patients with KRAS-mutated tumours. Thus, the effect of PIK3CA mutation may be potentially limited to patients with KRAS wild-type tumours[ 81 ]. Following the fact that only patients with KRAS-wild type CRC may respond to anti-EGFR antibodies, several studies have investigated the role of PIK3CA mutations on CRC cells response to cetuximab or panitumumab. The data collected so far indicate that CRC with PIK3CA mutations are significantly resistant to anti-EGFR antibodies. When only KRAS wild-type tumours are analyzed, the correlation is even stronger[ 83 - 85 ]. Changes in PIK3 signalling and loss of PTEN expression have been generally linked with the lack of response to EGFR-targeted therapy[ 86 , 87 ]. Recent studies have found that inhibition of cyclooxygenase-2 by regular use of aspirin after CRC diagnosis was associated with longer cancer specific survival time among patients with mutated as opposed to wild-type PIK3CA. The authors conclude that PIK3CA mutations may serve as a predictive biomarker for adjuvant aspirin therapy[ 88 ]. Further studies involving KRAS mutated CRC patients are necessary to establish the role of aspirin in PI3K pathway.
Biomarker of the potential toxicity of irinotecan
Irinotecan is a chemotherapeutic agent that inhibits topoisomerase I, thereby inhibiting replication and stimulating cell apoptosis. Advanced neutropenia and intensive diarrhoea caused by damaged intestinal epithelium are the most common adverse effects of irinotecan, which significantly limit its use. UGT1A1 gene polymorphism is a very useful biomarker of the potential toxicity of irinotecan. It appears that the use of genetic tests is reasonable before treatment initiation with irinotecan to avoid severe adverse effects - mainly neutropenia in women. Genotyping for UGT1A1 can be carried out to select a group of sensitive patients with UGT1A1*28 allele, of whom lower initial doses would be recommended. Hopefully, it will also allow to administer a higher accumulated dose of the drug, divided into smaller portions, to limit its toxicity[ 89 , 90 ]. However, according to a recent meta-analysis, genotyping for UGT1A1 has no predictive value in terms of responsiveness to various doses of irinotecan among patients with CRC[ 91 ]. On the other is recommended by ESMO for patients with several toxicity reaction in whom irinotecan in high doses should be used[ 33 ]. Furthermore, homozygosity for the UGT1A1*28 has been linked with improved efficacy of FOLFIRI[ 92 ].
Potential biomarkers of vascular endothelial growth factor - targeted therapy
Since the vascular endothelial growth factor (VEGF) - targeted therapy has been integrated into CRC treatment protocols, some anti-angiogenic drugs have been introduced (bevacizumab, regorafenib, aflibercept). However, a patient selection strategy to identify those patients who benefit most from this therapy has yet to be developed. To date, a predictive biomarker for bevacizumab - the most commonly administered anti-angiogenic drug in CRC therapy - has not yet been identified. Several studies on the identification of predictive biomarkers of bevacizumab have been performed. Jürgensmeier et al[ 93 ] evaluated retrospectively, using samples from randomised trial HORIZON III, the prognostic/predictive value of VEGF and soluble VEGF receptor-2. High baseline values of VEGF were associated with worse progression free survival (PFS) and overall survival. These data have revealed that baseline VEGF levels were not predictive of PFS or OS outcome in bevacizumab-treated patients[ 93 ]. Other studies have demonstrated that plasma VEGF-A may serve as a prognostic marker, but is unable to predict response to VEGF-targeted therapy in advanced CRC[ 94 , 95 ]. At the same time, KRAS mutation was found to be of no use as a predictive factor for bevacizumab[ 75 ].
Ezrin protein, a part of ezrin/radixin/moesin family may play an important role in tumour invasion process. Recent studies has found that overexpression of ezrin protein correlates with CRC aggressiveness, its metastatic potential and worse prognosis. High ezrin expression was also identified as marker of early local recurrence of rectal cancer[ 96 , 97 ]. Although further investigation is needed, ezrin may represent a relevant biomarker and target for personalized anti-metastatic therapies.
The recent studies result in a better understanding of colorectal cancer and assist in the development of new treatment regimens, especially in advanced CRC stages. The new predictive factors, molecular imaging, or even commercial genome tests increasingly facilitate tumour genome testing and assist in selecting targeted therapies. Adjuvant targeted therapy with anti-EGFR antibodies is required in advanced CRC patients and absence of KRAS , BRAF , NRAS and PIK3CA genes mutation. Tests for MSI or MSS tumour phenotype and the presence or absence of 18q chromosome deletion is very much desirable in standard therapy based on 5-FU. Genotyping of UGT1A1 alleles is reasonable before treatment initiation with irinotecan to avoid severe adverse effects. Further studies are necessary to identify predictive biomarker of bevacizumab. Targeted therapy against membrane receptors appears to be the future of CRC therapy. Some promising studies are now carried out in this area, dedicated to, inter alia, other EGFR ligands, insulin-like growth factor receptor 1, platelet-derived growth factor receptors and c-MET inhibitors. The aim of future research is to identify those biomarkers that can provide a non-invasive and cost-effective diagnosis, as well as to recognise the best prognostic panel of biomarkers and define the predictive biomarkers for available treatments.
Conflict-of-interest statement: The authors declare no conflicts of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: May 8, 2015
First decision: September 9, 2015
Article in press: December 21, 2015
P- Reviewer: Bajenova O S- Editor: Gong ZM L- Editor: A E- Editor: Wang CH

COMMENTS
Tumor biomarkers, the substances which are produced by tumors or the body's responses to tumors during tumorigenesis and progression, have been demonstrated to possess critical and encouraging ...
Tumor biomarkers are substances synthesized by cancer cells or other body cells in response to cancer and released into the circulation.[1] Tumor biomarkers vary widely in structure and may be simple molecules, such as catecholamines; well-characterized proteins, such as hormones, enzymes, or gene products; or more heterogeneous glycoproteins or mucins, such as carbohydrate antigen 125 (CA 125 ...
The initial markers tested for tumor staging were circulating DNA, as shown in Figure 5. Elevated concentrations of serum DNA have been linked to cancer (most particularly, metastatic cancer). Oncogene alterations, mismatch-repair gene mutations, and mutations in tumor suppressor genes can all be used as DNA biomarkers.
An ideal tumor marker should be highly sensitive, specific, reliable with high prognostic value, organ specificity and it should correlate with tumor stages. ... Bone. Ph.D. thesis, Osmania ...
Tumor Markers comprise a wide spectrum of biomacromolecules synthesized in excess concentration by a wide variety of neoplastic cells. The markers could be endogenous products of highly active metabolic malignant cells or the products of newly switched on genes, which remained unexprssed in early life or newly acquired antigens at cellular and sub-cellular levels.
Quantitative alterations in serum tumor markers have been expanded as noninvasive instrumentation to assess the effectiveness of treatment in human malignancies . In the BC, three serum tumor marker tumors, CEA, CA-15-3, and CA125, are the most widely used serum tumor markers in the clinical field .
Abstract. Tumor markers are playing an increasingly important role in cancer detection and management. These laboratory-based tests are potentially useful in screening for early malignancy, aiding cancer diagnosis, determining prognosis, surveillance following curative surgery for cancer, up front predicting drug response or resistance, and monitoring therapy in advanced disease. Clinically ...
Tumor markers are playing an increasingly important role in cancer detection and management. These laboratory-based tests are potentially useful in screening for early malignancy, aiding cancer diagnosis, determining prognosis, surveillance following curative surgery for cancer, up front predicting drug response or resistance, and monitoring therapy in advanced disease.
The possible clinical uses of tumor markers are manifold, and several categories of markers can be defined. A diag-nostic tumor marker is a marker that will aid in detection of malignant disease in an individual. Preferably, the marker should be tissue specific and not be influenced by benign diseases of the particular tissue/organ. Thus, a ...
Abstract. Due to the fact that the use of prostate-specific antigen (PSA) in prostate cancer (PC) management is limited, the search for novel biomarkers useful for early detection and monitoring, especially from noninvasive biofluids is an important pursuit. Urine has become one of the most attractive biofluids in clinical practice.
ent tumor markers inside and outside the cells (Fig. 1). The potential role of different assays in early tumor diagnosis and the challenges they still face are also dis-cussed. Overall, this paper ...
Biomarkers are helpful for disease diagnosis, monitoring disease progression, predicting disease recurrence, treatment monitoring, and efficacy, especially in the domain of cancer management and therapeutics. Clinical cancer biomarker utilization is progressively increasing with increase in and better access to healthcare by the growing populations worldwide. Early cancer detection and ...
Introduction. Colorectal cancer (CRC) is the third most commonly diagnosed cancer among adults and is the third leading cause of cancer-related death in the United States ().Most colorectal cancers occur sporadically and are characterized by a sequenced carcinogenesis process that involves the progressive accumulation of mutations in a period that lasts on average 10-15 years (2-5).
Routine measurement of tumor markers is not recommended in daily clinical practice for patients with cancer of unknown primary (CUP). We evaluated the diagnostic value of tumor markers in identifying favorable or unfavorable subsets in patients with CUP. We retrospectively reviewed the medical records of patients who were diagnosed with CUP between October 2010 and July 2015 at the National ...
IDEAL TUMOR MARKER. Only a few tumor markers have stood the test of time and entered in the diagnostic or management algorithms for clinicians.[1,2,5]The three most important characteristics of an ideal tumor marker are (a) it should be highly specific to a given tumor type, (b) it should provide a lead-time over clinical diagnosis and (c) it should be highly sensitive to avoid false positive ...
Tumor markers have traditionally been proteins or other substances that are made at higher amounts by cancer cells than normal cells. These can be found in the blood, urine, stool, tumors, or other tissues or bodily fluids of some patients with cancer. Increasingly, however, genomic markers (such as tumor gene mutations, patterns of tumor gene ...
A tumor marker is any substance in your body that provides information about a cancer. Traditionally, tumor markers refer to proteins that cancer cells make. Noncancerous cells can also make tumor markers in response to a cancer. But a tumor marker can also refer to mutations (changes) or patterns in your DNA.
Tumor markers may also be called cancer markers or biomarkers. A tumor marker is a substance found in the blood, urine, stool, the tumor itself, or tissue. The level of the tumor marker and which one it is can help you and your provider learn about your cancer. Some of these markers are specific to one cancer and others are seen in many types ...
More recently, tumor markers are identified as certain proteins or genes expressed in the tumor itself. Some of these markers expressed are prognostic in nature and are therefore important in predicting the malignancy of CRC-related tumor. While the predictive markers are often targeted for treatment, others serve diagnostic purposes .
Within the marker profile, the leading markers suggest the most probable histologies, as follows: in adenocarcinoma CEA; in squamous cell carcinoma CYFRA 21-1 and SCC; in large cell cancer CYFRA 21-1 and NSE; and in small cell lung cancer NSE and ProGRP. Most markers including CYFRA 21-1, CEA, NSE and SCC correlate clearly with tumor burden.
Besides tissue-based markers, ctDNA, proteomic, epigenomic, and metabolomic markers have recently enlarged this constantly evolving panorama. In particular, the liquid biopsy, based on the detection of tumor-related biomarkers in body fluids such as peripheral blood, has emerged as a very promising and noninvasive diagnostic tool that can ...
These tests look for tumor markers, which are sometimes called cancer markers. Tumor markers are substances that are often made by cancer cells or by normal cells in response to cancer. For example, some tumor markers are proteins that certain cancer cells make in larger amounts than normal cells do. Changes in the genes and other parts of ...
Abstract. Colorectal cancer (CRC) is the second most commonly diagnosed cancer among females and third among males worldwide. It also contributes significantly to cancer-related deaths, despite the continuous progress in diagnostic and therapeutic methods. Biomarkers currently play an important role in the detection and treatment of patients ...